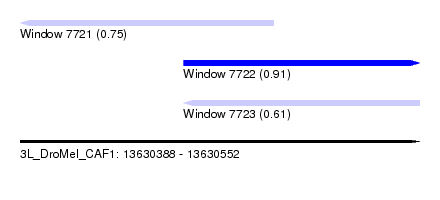
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,630,388 – 13,630,552 |
| Length | 164 |
| Max. P | 0.909583 |

| Location | 13,630,388 – 13,630,492 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 75.99 |
| Mean single sequence MFE | -24.93 |
| Consensus MFE | -16.15 |
| Energy contribution | -18.14 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.751001 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13630388 104 - 23771897 AAAAAGGCAACCACCGCAUGGCUACAACUACUGCUGC------UGAUUCUGCUGCCAACUAUGAGCCAGGGGAAGUCCCCAGUCUAC-CCAGUUGCCCUUUCCUUUCCUGC .....((((((.......(((((.((......((.((------.......)).))......)))))))((((....)))).......-...)))))).............. ( -28.90) >DroSec_CAF1 37862 92 - 1 AAAAAGGCAACCACCGCAUGGCUACAACUACUGC------------------UGCCAACUAUGAGCCAGGGGAAGUCCCCAGUCUCC-UCAGUUGCCCUGUCCCUUCCUUC .....((((((.......((((..((.....)).------------------.)))).....(((.(.((((....)))).).))).-...)))))).............. ( -25.60) >DroSim_CAF1 47678 92 - 1 AAAAAGGCAACCACCGCAUGGCUACAACUACUGC------------------UGCCAACUAUGAGCCAGGGGAAGUCCCCAGUCUCC-UCAGUUGCCCUGUCCCUUCCUUC .....((((((.......((((..((.....)).------------------.)))).....(((.(.((((....)))).).))).-...)))))).............. ( -25.60) >DroEre_CAF1 40176 93 - 1 AAAAAGGCAACCACCGCACGACUACAACUACUGCUGCUGCUUUUGCUUCUGCUGCCAACUAUGAGCCAGGGGAAGUCCCCAGUCUCCCCCCCA------------------ .....((((......(((.............))).((.((....))....))))))......(((.(.((((....)))).).))).......------------------ ( -19.62) >consensus AAAAAGGCAACCACCGCAUGGCUACAACUACUGC__________________UGCCAACUAUGAGCCAGGGGAAGUCCCCAGUCUCC_CCAGUUGCCCUGUCCCUUCCUUC .....((((((........((((.((......((.((.............)).))......)))))).((((....))))...........)))))).............. (-16.15 = -18.14 + 2.00)

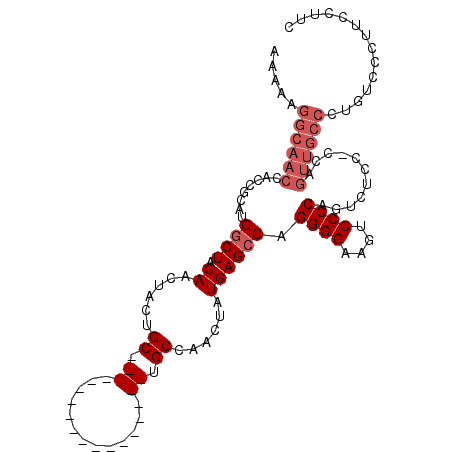
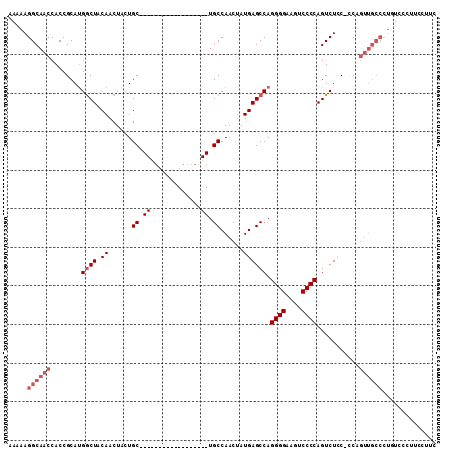
| Location | 13,630,455 – 13,630,552 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.31 |
| Mean single sequence MFE | -25.32 |
| Consensus MFE | -16.84 |
| Energy contribution | -16.73 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.67 |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.909583 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13630455 97 + 23771897 ---GCAGCAGUAGUUGUAGCCAUGCGGUGGUUGCCUUUUUGUGCCCAUGCGCAUGGAAAAUAGUUGCACAAACUUGCA------CAGAGCAAAAAAAAAAAG-------------GA-GG ---...((((..(((....(((((((((((..((......))..)))).)))))))..)))..))))......((((.------....))))..........-------------..-.. ( -26.60) >DroPse_CAF1 21528 80 + 1 ----------------UAGUCG--UGGUGGUUGCCUUUUUGUGCCCAUGCGCAUGGAAAAUAGUUGCACAAACUUGCA------CACAGCACACAAA--AAG-------------AU-AG ----------------..((.(--((.((..(((...(((((((((((....)))).........)))))))...)))------..)).)))))...--...-------------..-.. ( -20.70) >DroSec_CAF1 37920 93 + 1 ------GCAGUAGUUGUAGCCAUGCGGUGGUUGCCUUUUUGUGCCCAUGCGCAUGGAAAAUAGUUGCACAAACUUGCA------CAGAGCAAAAA-AAAAAG-------------AA-GG ------((((..(((....(((((((((((..((......))..)))).)))))))..)))..))))......((((.------....))))...-......-------------..-.. ( -26.40) >DroSim_CAF1 47736 94 + 1 ------GCAGUAGUUGUAGCCAUGCGGUGGUUGCCUUUUUGUGCCCAUGCGCAUGGAAAAUAGUUGCACAAACUUGCA------CAGAGCAAAAAAAAAAAG-------------AA-GG ------((((..(((....(((((((((((..((......))..)))).)))))))..)))..))))......((((.------....))))..........-------------..-.. ( -26.40) >DroEre_CAF1 40229 120 + 1 GCAGCAGCAGUAGUUGUAGUCGUGCGGUGGUUGCCUUUUUGUGCCCAUGCGCAUGGAAAAUAGUUGCACAAACUUGCACAGGCACAGAGCAAAAAAGAAAAAAAAAAAAAGAAUAAACGG ((.(((((....))))).((((((((((....)))..(((((((((((....)))).........)))))))...)))).))).....)).............................. ( -27.30) >DroAna_CAF1 74056 76 + 1 -----------------AGCCGAGCGGUGGUUGCCUUUUUGUGCCCAUGCGCAUGGAAAAUAGUGGCACAAACUUGCA------CGG--CAAAAA-GAUAAA------------------ -----------------.((((.(((((...((((((((..(((......)))..)))).....))))...)).))).------)))--).....-......------------------ ( -24.50) >consensus ______GCAGUAGUUGUAGCCAUGCGGUGGUUGCCUUUUUGUGCCCAUGCGCAUGGAAAAUAGUUGCACAAACUUGCA______CAGAGCAAAAAAAAAAAG_____________AA_GG ...................(((((((((((..((......))..)))).))))))).........(((......)))........................................... (-16.84 = -16.73 + -0.11)

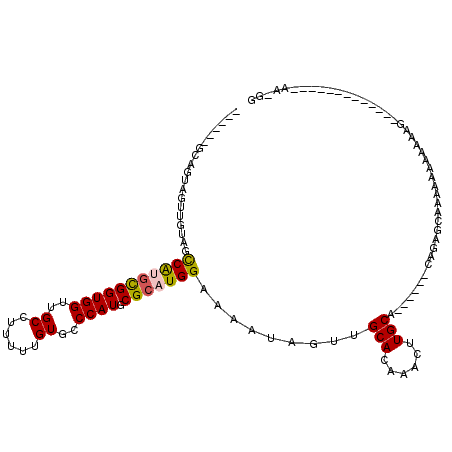
| Location | 13,630,455 – 13,630,552 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.31 |
| Mean single sequence MFE | -20.62 |
| Consensus MFE | -14.23 |
| Energy contribution | -14.23 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.06 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.608427 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13630455 97 - 23771897 CC-UC-------------CUUUUUUUUUUUGCUCUG------UGCAAGUUUGUGCAACUAUUUUCCAUGCGCAUGGGCACAAAAAGGCAACCACCGCAUGGCUACAACUACUGCUGC--- ..-.(-------------((((((.....(((((((------((((((((.....))))........)))))).))))).)))))))........((((((......))).)))...--- ( -20.10) >DroPse_CAF1 21528 80 - 1 CU-AU-------------CUU--UUUGUGUGCUGUG------UGCAAGUUUGUGCAACUAUUUUCCAUGCGCAUGGGCACAAAAAGGCAACCACCA--CGACUA---------------- ..-.(-------------(((--(((((((.(((((------((((((((.....))))........)))))))))))))))))))).........--......---------------- ( -22.80) >DroSec_CAF1 37920 93 - 1 CC-UU-------------CUUUUU-UUUUUGCUCUG------UGCAAGUUUGUGCAACUAUUUUCCAUGCGCAUGGGCACAAAAAGGCAACCACCGCAUGGCUACAACUACUGC------ ..-..-------------......-...(((((.((------(((.....((((((...........))))))...)))))....))))).....((((((......))).)))------ ( -18.70) >DroSim_CAF1 47736 94 - 1 CC-UU-------------CUUUUUUUUUUUGCUCUG------UGCAAGUUUGUGCAACUAUUUUCCAUGCGCAUGGGCACAAAAAGGCAACCACCGCAUGGCUACAACUACUGC------ ..-..-------------..........(((((.((------(((.....((((((...........))))))...)))))....))))).....((((((......))).)))------ ( -18.70) >DroEre_CAF1 40229 120 - 1 CCGUUUAUUCUUUUUUUUUUUUUCUUUUUUGCUCUGUGCCUGUGCAAGUUUGUGCAACUAUUUUCCAUGCGCAUGGGCACAAAAAGGCAACCACCGCACGACUACAACUACUGCUGCUGC .(((...................((((((((((((((((.((((((......)))).........)).))))).))))).))))))((.......))))).................... ( -21.20) >DroAna_CAF1 74056 76 - 1 ------------------UUUAUC-UUUUUG--CCG------UGCAAGUUUGUGCCACUAUUUUCCAUGCGCAUGGGCACAAAAAGGCAACCACCGCUCGGCU----------------- ------------------......-.....(--(((------.((...((((((((.(................)))))))))..((......)))).)))).----------------- ( -22.19) >consensus CC_UU_____________CUUUUUUUUUUUGCUCUG______UGCAAGUUUGUGCAACUAUUUUCCAUGCGCAUGGGCACAAAAAGGCAACCACCGCACGGCUACAACUACUGC______ ..........................................(((...(((((((.........((((....)))))))))))...)))............................... (-14.23 = -14.23 + -0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:36:17 2006