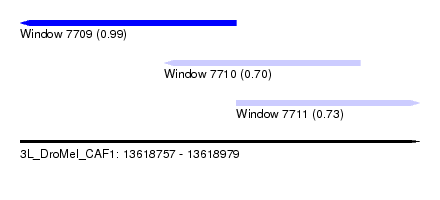
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,618,757 – 13,618,979 |
| Length | 222 |
| Max. P | 0.987161 |

| Location | 13,618,757 – 13,618,877 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.12 |
| Mean single sequence MFE | -30.68 |
| Consensus MFE | -28.60 |
| Energy contribution | -28.80 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.93 |
| SVM decision value | 2.07 |
| SVM RNA-class probability | 0.987161 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13618757 120 - 23771897 AUUCGGAUUCAGGUUCAAAUUCAGAUUCCUGACUUGGCAAUAUUUCAAUCAAACUUUUUUUUUGCCACUCUACGGGGCGGCUGUGUAGCCGUCCAAAUGGAUAUUUAUGCUUAUCAUGCU ...((...(((((.((.......))..)))))..((((((.....................)))))).....))((((((((....)))))))).....((((........))))..... ( -29.70) >DroSec_CAF1 25706 117 - 1 AUUCGGAUUCAGGUUCGAAUUCAGAUUCCUGACUUGGCAAUAUUUCAAUCAAACUUU---UUUGCCACUCUACGGGGCGGCUGUGUAGCCGUCCAAAUGGAUAUUUAUGCUUAUCAUGCU ((((((((....))))))))((((....))))..((((((.................---.)))))).((((..((((((((....))))))))...)))).......((.......)). ( -31.87) >DroSim_CAF1 34774 117 - 1 AUUCGGAUUCAGGUUCGAAUUCAGAUUCCUGACUUGGCAAUAUUUCAAUCAAACUUU---UUUGCCACUCUACGGGGCGGCUGUGUAGCCGUCCAAAUGGAUAUUUAUGCUUAUCAUGCU ((((((((....))))))))((((....))))..((((((.................---.)))))).((((..((((((((....))))))))...)))).......((.......)). ( -31.87) >DroEre_CAF1 28483 117 - 1 AUUCGGAUUCAGGCUCAAAUUCAGAUUCCUGACUUGGCAAUAUUUCAAUCAAACUUU---UUUGCCACUCUACGGGGCGGCUGUGUAGCCGUCCAAAUGGAUAUUUAUGCUUAUCAUGCU .....(((..((((..((((((((....))))..((((((.................---.)))))).((((..((((((((....))))))))...)))).))))..)))))))..... ( -29.57) >DroYak_CAF1 32805 117 - 1 AUAAGGAUUCAGGUUGAAAUUCAGAUUCCUGACUUGGCAAUAUUUCAAUCAAACUUU---UUUGCCACUCUACGGGGCGGCUGUGUAGCCGUCCAAAUGGAUAUUUAUGCUUAUCAUGCU (((((.((((((((((.....)))...)))))..((((((.................---.)))))).((((..((((((((....))))))))...)))).....)).)))))...... ( -30.37) >consensus AUUCGGAUUCAGGUUCAAAUUCAGAUUCCUGACUUGGCAAUAUUUCAAUCAAACUUU___UUUGCCACUCUACGGGGCGGCUGUGUAGCCGUCCAAAUGGAUAUUUAUGCUUAUCAUGCU ........(((((.((.......))..)))))..((((((.....................)))))).((((..((((((((....))))))))...)))).......((.......)). (-28.60 = -28.80 + 0.20)

| Location | 13,618,837 – 13,618,946 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 86.26 |
| Mean single sequence MFE | -32.84 |
| Consensus MFE | -24.44 |
| Energy contribution | -25.40 |
| Covariance contribution | 0.96 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.701616 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13618837 109 - 23771897 AAUCAAUUAAAAUACGUGCCGGGCAUUAGAAGAGUGCGGCGCAUAUACGGC-CGGAGGAUCCGGCUACGGAUUCGGAUUCAGGUUCAAAUUCAGAUUCCUGACUUGGCAA ...............((((((.((.........)).)))))).......((-(((.(((((((....)))))))....(((((.((.......))..))))).))))).. ( -36.50) >DroSec_CAF1 25783 110 - 1 AAUCAAUUAAAAUACGUGCCGGGCAUUAGAAGAGUGCGGCGCAUGUACGGCCCUGAGGAUCCGGCUACGGAUUCGGAUUCAGGUUCGAAUUCAGAUUCCUGACUUGGCAA ...............((((((.((.........)).)))))).(((.((..((((((((((((....)))))))....)))))..))...((((....))))....))). ( -37.60) >DroSim_CAF1 34851 110 - 1 AAUCAAUUAAAAUACGUGCCGGGCAUUAGAAGAGUGCGGCGCAUGUACGGCCCUGAGGAUCCGGCUACGGAUUCGGAUUCAGGUUCGAAUUCAGAUUCCUGACUUGGCAA ...............((((((.((.........)).)))))).(((.((..((((((((((((....)))))))....)))))..))...((((....))))....))). ( -37.60) >DroEre_CAF1 28560 104 - 1 AAUCAAUUAAAAUACGUGCCGGGCAUUAGAAGAGUGCGGCGCAUGGA------UUCGUAUUCGUAUUCGUAUUCGGAUUCAGGCUCAAAUUCAGAUUCCUGACUUGGCAA ...............((((((.((.........)).)))))).((((------((((....((....))....)))))))).((.(((..((((....)))).))))).. ( -29.20) >DroYak_CAF1 32882 93 - 1 AAUCAAUUAAAAUACGUGCCGGGCAUUAGAAGAGUGCGGCGCAU--------------AUGCG---GCAUAUAAGGAUUCAGGUUGAAAUUCAGAUUCCUGACUUGGCAA ................(((((((..........((((.(((...--------------.))).---))))........((((((((.....)))...)))))))))))). ( -23.30) >consensus AAUCAAUUAAAAUACGUGCCGGGCAUUAGAAGAGUGCGGCGCAUGUACGGC_CUGAGGAUCCGGCUACGGAUUCGGAUUCAGGUUCAAAUUCAGAUUCCUGACUUGGCAA .(((...........((((((.((.........)).))))))..............(((((((....))))))).)))(((((.((.......))..)))))........ (-24.44 = -25.40 + 0.96)

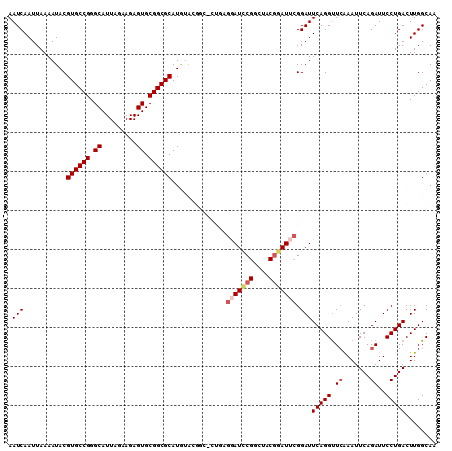
| Location | 13,618,877 – 13,618,979 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 78.40 |
| Mean single sequence MFE | -32.85 |
| Consensus MFE | -23.10 |
| Energy contribution | -22.72 |
| Covariance contribution | -0.39 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.730166 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13618877 102 + 23771897 CCGUAGCCGGAUCCUCCG-GCCGUAUAUGCGCCGCAC---UCUUCUAAUGCCCGGCACGUAUUUUAAUUGAUUCGUGCCGCACAAGGAGCCGCAGGAGUGGCAU-CA---- .....(((((.....)))-)).........(((((.(---((((((..(((..((((((.(((......))).)))))))))..))))).....)).)))))..-..---- ( -37.00) >DroPse_CAF1 8628 100 + 1 --CGAGCCGCCUGGCCACGACCA---------CACACUCUUCUUCUAAUGCUUGGCACGUAUUUUAAUUGAUUCGUGCCACACAGGGAGCGGCACCAGCAGCAGGCAGUAC --...(((((((((((..((...---------.....))..(((((..((..(((((((.(((......))).)))))))..))))))).)))..)))..)).)))..... ( -28.20) >DroSec_CAF1 25823 103 + 1 CCGUAGCCGGAUCCUCAGGGCCGUACAUGCGCCGCAC---UCUUCUAAUGCCCGGCACGUAUUUUAAUUGAUUCGUGCCGCACAAGGAGCCGCAGGAGUGGCAU-CA---- .....((((..((((...(((.((....)))))((..---.(((((..(((..((((((.(((......))).)))))))))..)))))..)))))).))))..-..---- ( -35.60) >DroSim_CAF1 34891 103 + 1 CCGUAGCCGGAUCCUCAGGGCCGUACAUGCGCCGCAC---UCUUCUAAUGCCCGGCACGUAUUUUAAUUGAUUCGUGCCGCACAAGGAGCCGCAGGAGUGGCAU-CA---- .....((((..((((...(((.((....)))))((..---.(((((..(((..((((((.(((......))).)))))))))..)))))..)))))).))))..-..---- ( -35.60) >DroEre_CAF1 28600 97 + 1 ACGAAUACGAAUACGAA------UCCAUGCGCCGCAC---UCUUCUAAUGCCCGGCACGUAUUUUAAUUGAUUCGUGCCGCACAAGGAGCCGCAGGAGUGGCAU-CA---- ..............((.------(((.((.((.(((.---........)))..((((((.(((......))).)))))))).)).)))(((((....))))).)-).---- ( -28.80) >DroYak_CAF1 32922 86 + 1 AUGC---CGCAU--------------AUGCGCCGCAC---UCUUCUAAUGCCCGGCACGUAUUUUAAUUGAUUCGUGCCGCACAAGGAGCCGCAGGAGUGGCAU-CA---- ((((---(((..--------------.((((.....(---((((....((..(((((((.(((......))).)))))))..))))))).))))...)))))))-..---- ( -31.90) >consensus CCGUAGCCGGAUCCUCAG_GCCGUACAUGCGCCGCAC___UCUUCUAAUGCCCGGCACGUAUUUUAAUUGAUUCGUGCCGCACAAGGAGCCGCAGGAGUGGCAU_CA____ ..........................................((((..((..(((((((.(((......))).)))))))..))))))(((((....)))))......... (-23.10 = -22.72 + -0.39)

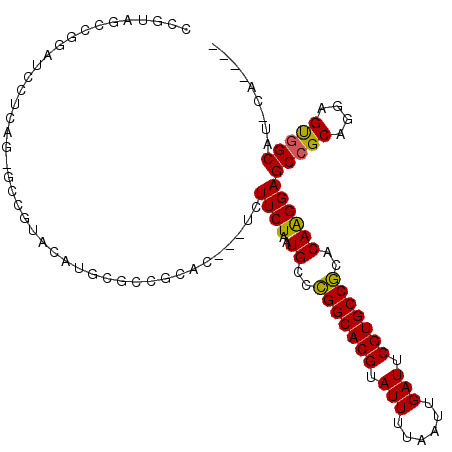
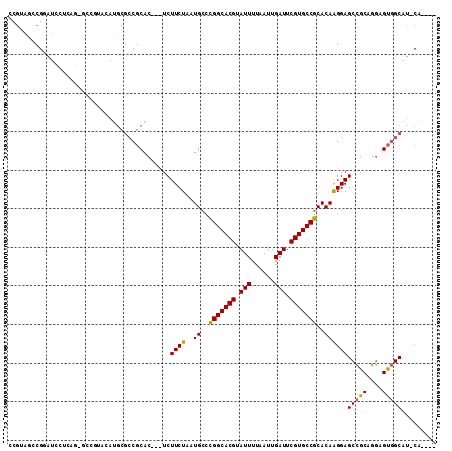
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:36:06 2006