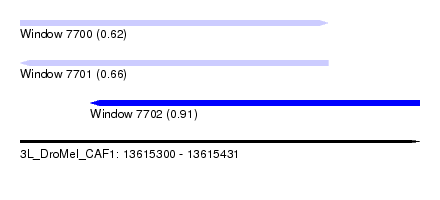
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,615,300 – 13,615,431 |
| Length | 131 |
| Max. P | 0.909142 |

| Location | 13,615,300 – 13,615,401 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 76.96 |
| Mean single sequence MFE | -40.23 |
| Consensus MFE | -22.97 |
| Energy contribution | -23.33 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.619466 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13615300 101 + 23771897 CCCCC--CA-----------UGCCAGCGUUGUUGUGGGUGGUCCUUUUUGGGCGG--GGGGCUAUGAUGGGUGCAUGGACCCAUAAACUGCGUUAGUUUAUGAUGCCCAAGCGCUA (((((--(.-----------.(((.(((....))).))).((((.....))))))--))))........(((((.(((.(.(((((((((...)))))))))..).))).))))). ( -41.30) >DroSec_CAF1 22296 100 + 1 CCCCCCCCU-----------AGCUAGCGUUGUUGUGGGUGGUCCUUUUUGG---G--GGGGCUGUGCUGGGUGCAUGGACCCAUAAACUGCGUUAGUUUAUGAUGCCCGAGCGCUA .((((((((-----------(((.......)))).(((....)))....))---)--))))..(((((.((.((((.....(((((((((...))))))))))))))).))))).. ( -40.50) >DroSim_CAF1 31390 99 + 1 CCCCCCUCU-----------UGCCAGCGUUGUUGUGGGUGGUCCUUUUCGG------GGGGCUGUGCUGGGUGCAUGGACCCAUAAACUGCGUUAGUUUAUGAUGCCCGAGCGCUA ((((((...-----------.(((.(((....))).)))((......))))------))))..(((((.((.((((.....(((((((((...))))))))))))))).))))).. ( -39.10) >DroEre_CAF1 25116 112 + 1 CUCCCCCCAUUCUCCC--CAUUCU--CGCUGUUGUGGGAGGUCCUUUUUGGGGGGCGGGGGCUGUGCUGGGUGCAUGGACCCAUAAACUGCGUUAGUUUAUGAUGCCCGAGCGCUA ...(((((...(((((--((((((--(((....)))))))........))))))).)))))..(((((.((.((((.....(((((((((...))))))))))))))).))))).. ( -49.90) >DroYak_CAF1 29426 116 + 1 CCCCUCCCCCACCGCCGGCUUUCCUGCGUUGUUGUGGGAGGUCCUUUUUGGGGAGUUGGGGCUUUGCUGGCUGCAUGGACCCAUAAACUGCGUUAGUUUAUGAUGCCCGAGCGCUA ((((.((((((((((.((....)).))).....))))).))........)))).(((.((((..(((.....)))......(((((((((...)))))))))..)))).))).... ( -41.90) >DroAna_CAF1 58694 89 + 1 CUCCC----------------GGCAUCUACGAUGUGGCA-GUCA----------GCAUGGAGUGUCCUGGGUGCAUGGACCCAUAAACUCCGUUAGUUUAUGAUGCCCGAGCGCUA (((..----------------((((((.(((((......-))).----------..(((((((....(((((......)))))...)))))))..))....)))))).)))..... ( -28.70) >consensus CCCCCCCCA___________UGCCAGCGUUGUUGUGGGAGGUCCUUUUUGG___G__GGGGCUGUGCUGGGUGCAUGGACCCAUAAACUGCGUUAGUUUAUGAUGCCCGAGCGCUA .((((................(((..((....))..)))...((.....))......))))..(((((.((.((((.....((((((((.....)))))))))))))).))))).. (-22.97 = -23.33 + 0.36)



| Location | 13,615,300 – 13,615,401 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 76.96 |
| Mean single sequence MFE | -34.87 |
| Consensus MFE | -18.02 |
| Energy contribution | -18.63 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.658139 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13615300 101 - 23771897 UAGCGCUUGGGCAUCAUAAACUAACGCAGUUUAUGGGUCCAUGCACCCAUCAUAGCCCC--CCGCCCAAAAAGGACCACCCACAACAACGCUGGCA-----------UG--GGGGG ....((.(((((.(((((((((.....)))))))))))))).))...........((((--(((.((.....)).....((((......).)))..-----------))--))))) ( -33.80) >DroSec_CAF1 22296 100 - 1 UAGCGCUCGGGCAUCAUAAACUAACGCAGUUUAUGGGUCCAUGCACCCAGCACAGCCCC--C---CCAAAAAGGACCACCCACAACAACGCUAGCU-----------AGGGGGGGG ....((..((((.(((((((((.....)))))))))))))..))...........((((--(---((.....((.....))........(....).-----------.))))))). ( -35.00) >DroSim_CAF1 31390 99 - 1 UAGCGCUCGGGCAUCAUAAACUAACGCAGUUUAUGGGUCCAUGCACCCAGCACAGCCCC------CCGAAAAGGACCACCCACAACAACGCUGGCA-----------AGAGGGGGG ....((..((((.(((((((((.....)))))))))))))..))...........((((------((.....((.....)).........((....-----------)).)))))) ( -31.30) >DroEre_CAF1 25116 112 - 1 UAGCGCUCGGGCAUCAUAAACUAACGCAGUUUAUGGGUCCAUGCACCCAGCACAGCCCCCGCCCCCCAAAAAGGACCUCCCACAACAGCG--AGAAUG--GGGAGAAUGGGGGGAG ....(((.((((.(((((((((.....))))))))))))).(((.....))).))).....(((((((........(((((.((......--....))--)))))..))))))).. ( -38.20) >DroYak_CAF1 29426 116 - 1 UAGCGCUCGGGCAUCAUAAACUAACGCAGUUUAUGGGUCCAUGCAGCCAGCAAAGCCCCAACUCCCCAAAAAGGACCUCCCACAACAACGCAGGAAAGCCGGCGGUGGGGGAGGGG ..((((..((((.(((((((((.....)))))))))))))..)).))........((((...(((.......))).(((((((.....(((.((....)).)))))))))).)))) ( -42.60) >DroAna_CAF1 58694 89 - 1 UAGCGCUCGGGCAUCAUAAACUAACGGAGUUUAUGGGUCCAUGCACCCAGGACACUCCAUGC----------UGAC-UGCCACAUCGUAGAUGCC----------------GGGAG ....((..((((.(((((((((.....)))))))))))))..)).(((.((.....))..((----------...(-(((......))))..)).----------------))).. ( -28.30) >consensus UAGCGCUCGGGCAUCAUAAACUAACGCAGUUUAUGGGUCCAUGCACCCAGCACAGCCCC__C___CCAAAAAGGACCACCCACAACAACGCUGGCA___________AGGGGGGGG ((((((..((((.(((((((((.....)))))))))))))..))............................((.....))........))))....................... (-18.02 = -18.63 + 0.61)



| Location | 13,615,323 – 13,615,431 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.04 |
| Mean single sequence MFE | -30.68 |
| Consensus MFE | -21.30 |
| Energy contribution | -21.63 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.909142 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13615323 108 - 23771897 ----------UGUCGCUCUGCGUAUUUCGCUUUGCUCACUUAGCGCUUGGGCAUCAUAAACUAACGCAGUUUAUGGGUCCAUGCACCCAUCAUAGCCCC--CCGCCCAAAAAGGACCACC ----------.(((((...(((.....)))...)).......(((...((((...(((((((.....)))))))((((......))))......)))).--.)))........))).... ( -29.20) >DroPse_CAF1 4660 106 - 1 AUCCGCUCGCUGU--CUCAGCGCACUUCGCUUUGCUCACUUAGCGCUCGGGCAUCAUAAACUAACGCAGUUUAUGGGUCCAUGCACCCAAAGCAGAC------------AGGGAACGACA ...((.((.((((--((..((.......(((..(....)..)))((..((((.(((((((((.....)))))))))))))..)).......))))))------------)).)).))... ( -34.80) >DroSim_CAF1 31415 104 - 1 ----------UGUCGCUCUGCGUAUUUCGCUUUGCUCACUUAGCGCUCGGGCAUCAUAAACUAACGCAGUUUAUGGGUCCAUGCACCCAGCACAGCCCC------CCGAAAAGGACCACC ----------.(((((...(((.....)))...))...(((..((...((((.(((((((((.....))))))))).....(((.....)))..)))).------.))..)))))).... ( -29.10) >DroYak_CAF1 29462 110 - 1 ----------UGUCGCUCUGCGUAUUUCGCUUUGCUCACUUAGCGCUCGGGCAUCAUAAACUAACGCAGUUUAUGGGUCCAUGCAGCCAGCAAAGCCCCAACUCCCCAAAAAGGACCUCC ----------.(((((...)).......((((((((......((((..((((.(((((((((.....)))))))))))))..)).)).)))))))).................))).... ( -29.80) >DroAna_CAF1 58714 104 - 1 ----GAUUGCUGUC-CUCUGCGAUUUUCGCUUUGCUCACUUAGCGCUCGGGCAUCAUAAACUAACGGAGUUUAUGGGUCCAUGCACCCAGGACACUCCAUGC----------UGAC-UGC ----......((((-((..(((.....)))...(((.....)))((..((((.(((((((((.....)))))))))))))..))....))))))........----------....-... ( -29.50) >DroPer_CAF1 4642 106 - 1 AUCCGCUAGCUGU--CUCAGCGCACUUCGCUUUGCUCACUUAGCGCUCGGGCAUCAUAAACUAACGCAGUUUAUGGGUCCAUGCACCCAAAGCAGAC------------AGGGAACGACA .(((((..((((.--..)))))).....((((((((.....)))((..((((.(((((((((.....)))))))))))))..))....)))))....------------..)))...... ( -31.70) >consensus __________UGUC_CUCUGCGUAUUUCGCUUUGCUCACUUAGCGCUCGGGCAUCAUAAACUAACGCAGUUUAUGGGUCCAUGCACCCAGCACAGCCCC__________AAAGGACCACC ...........(((.....(((.....)))...(((.....)))((..((((.(((((((((.....)))))))))))))..)).............................))).... (-21.30 = -21.63 + 0.33)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:35:57 2006