| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 1,369,565 – 1,369,663 |
| Length | 98 |
| Max. P | 0.883370 |

| Location | 1,369,565 – 1,369,663 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 71.98 |
| Mean single sequence MFE | -29.27 |
| Consensus MFE | -16.50 |
| Energy contribution | -17.00 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.883370 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
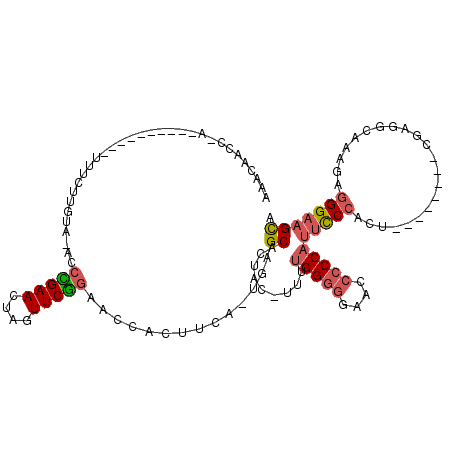
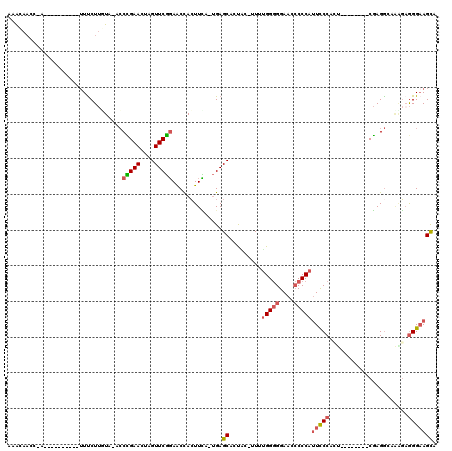
>3L_DroMel_CAF1 1369565 98 - 23771897 AAACAACC-A----------UUUCUUGUA-ACCUGAACUAGUUCGGAACCACUUUA-UGAGCACUGC-UUUUGGGGGAACCCCCAAUUCCCCC--------CGAGGCACAGAGGGAAGCA ........-.----------(..(((...-..(((((....)))))..........-.......(((-(((.(((((((.......)))))))--------.))))))..)))..).... ( -26.70) >DroSec_CAF1 25188 97 - 1 AAACAACC-A----------UUUCUUGUA-ACCCGAACUAGUUCGGAACCACUUCA-UGAGCACUAC-UUUUGG-GGAACACCCAUUCCCACU--------CGAGGCAAAGAGGGAAGCA ........-.----------.........-..(((((....)))))..........-...((.....-...(((-(.....))))(((((.((--------........)).))))))). ( -21.50) >DroSim_CAF1 22531 98 - 1 AAACAACC-A----------UUUCUUGUA-ACCCGAACUAGUUCGGAACCACUUCA-UGAGCACCAC-UUUUGGGGGAACCCCCAUUCCCACU--------CGAGGCAACGAGGGAAGCA ........-.----------(..(((((.-.(((((....((((((((....))).-))))).....-...(((((....))))).......)--------)).))..)))))..).... ( -27.20) >DroEre_CAF1 26231 110 - 1 ACGAAAACAUUUCCUGUAUGUUUCUUUUA-UACCGAACUAGUUCGGAACCACUUCU-UGAGCACUGCUUUUUGGGGGAAGCCCCAUUCCCAGU--------CGAGACUAGGAGGGCAGCA .........(((((((...(((((.....-..(((((....)))))..........-...(.((((.....(((((....)))))....))))--------)))))))))))))...... ( -30.00) >DroYak_CAF1 27418 109 - 1 AUAUAACC-AUUCCCGCACAAUUCUUUUA-UCCCGAACUAGUUCGGAACUACUUCA-UGAGCAUUUGCUUUUGGGGGAACCCCCAUUCCCAGU--------CGAGGCAAAGUGGGAAGCA ........-.(((((((............-..(((((....))))).....((((.-.((((....)))).(((((....)))))........--------.))))....)))))))... ( -33.80) >DroAna_CAF1 26382 107 - 1 -UACAUCC-U----------UUCCUAUUGCUCCGGAACUAGUUCUUAGCCAGUUGAGCGAGCAUCUC-GUUGGGGGGAAUCCCCAUUCCCGGCGCAUACUCCCAGGAAGGCAAGGAAGUA -(((.(((-(----------(.(((..(((.((((((((.((.....)).)))).((((((...)))-))).((((....))))....)))).)))...((....))))).))))).))) ( -36.40) >consensus AAACAACC_A__________UUUCUUGUA_ACCCGAACUAGUUCGGAACCACUUCA_UGAGCACUAC_UUUUGGGGGAACCCCCAUUCCCACU________CGAGGCAAAGAGGGAAGCA ................................(((((....)))))..............((.........(((((....)))))(((((......................))))))). (-16.50 = -17.00 + 0.50)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:43:12 2006