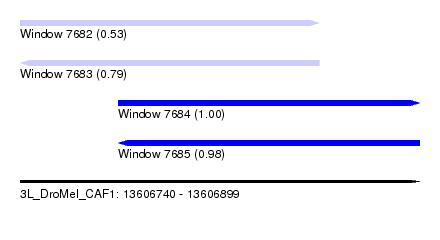
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,606,740 – 13,606,899 |
| Length | 159 |
| Max. P | 0.998601 |

| Location | 13,606,740 – 13,606,859 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.82 |
| Mean single sequence MFE | -23.77 |
| Consensus MFE | -20.36 |
| Energy contribution | -20.28 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.533704 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13606740 119 + 23771897 AC-UCGUAACUAUUACCAUAACAUUUAUGAUUAUUAACCUACAAUGCGGGGCACUCUAAAUGCAAUUAAGAUUACGAUCUAUAUUGCAUUUUCUGUGUUGCAAAAAGAAGCCUGGCAACU ..-((((((...............))))))..............(((.((((......((((((((..((((....))))..))))))))((((...........)))))))).)))... ( -23.86) >DroSec_CAF1 13925 119 + 1 AC-UCGUAACUAUUACCAUAACAUUUAUGAUUAUUAACCUACAAUGCGGGGCACUCUAAAUGCAAUUAAGAUUACGAUCUAUAUUGCAUUUUCUGUGUUGCAAAAAGAAGCCUGGCAACU ..-((((((...............))))))..............(((.((((......((((((((..((((....))))..))))))))((((...........)))))))).)))... ( -23.86) >DroSim_CAF1 22898 119 + 1 AC-UCGUAACUAUUACCAUAACAUUUAUGAUUAUUAACCUACAAUGCGGGGCACUCUAAAUGCAAUUAAGAUUACGAUCUAUAUUGCAUUUUCUGUGUUGCAAAAAGAAGCCUGGCUACU ..-..(((........((((.....))))................((.((((......((((((((..((((....))))..))))))))((((...........)))))))).))))). ( -22.90) >DroEre_CAF1 16526 119 + 1 AC-UCGUAACUAUUACCAUAACAUUUAUGAUUAUUAACCCACAAUGCGGGGCACUCUAAAUGCAAUUAAGAUUACGAUCUAUAUUGCAUUUUCUGUGUUGCAAAAAGAAGCCUGGCCACU ..-((((((...............))))))...............((.((((......((((((((..((((....))))..))))))))((((...........)))))))).)).... ( -21.96) >DroYak_CAF1 20719 119 + 1 AC-UCGUAACUAUUACCAUAACAUUUAUGAUUAUUAACCUACAAUGCGGGGCACUCUAAAUGCAAUUAAGAUUACGAUCUAUAUUGCAUUUUCUGUGUUGCAAAAAGGAGCCUGGCAACU .(-(((((.........((((((....)).))))..........))))))((((...(((((((((..((((....))))..)))))))))...))))(((....((....)).)))... ( -23.91) >DroAna_CAF1 49715 120 + 1 GCCCCGUAACUAUUACCGUAGCAUUUAUGAUUAUUAACCCACAAUGCGGGGCACUCUAAAUGCAAUUAAGAUUACGAUCUAUAUUACAUUUUCUGUGUUGCAAAAAGAAACCUGGCAACU ((((((((.((((....))))((....))...............))))))))((...(((((.(((..((((....))))..))).)))))...))(((((....((....)).))))). ( -26.10) >consensus AC_UCGUAACUAUUACCAUAACAUUUAUGAUUAUUAACCUACAAUGCGGGGCACUCUAAAUGCAAUUAAGAUUACGAUCUAUAUUGCAUUUUCUGUGUUGCAAAAAGAAGCCUGGCAACU ................(((((...)))))................((.((((......((((((((..((((....))))..))))))))((((...........)))))))).)).... (-20.36 = -20.28 + -0.08)


| Location | 13,606,740 – 13,606,859 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.82 |
| Mean single sequence MFE | -27.20 |
| Consensus MFE | -24.92 |
| Energy contribution | -25.17 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.20 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.793912 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13606740 119 - 23771897 AGUUGCCAGGCUUCUUUUUGCAACACAGAAAAUGCAAUAUAGAUCGUAAUCUUAAUUGCAUUUAGAGUGCCCCGCAUUGUAGGUUAAUAAUCAUAAAUGUUAUGGUAAUAGUUACGA-GU .((((((((((((((.((((.....))))(((((((((..((((....))))..))))))))))))).)))..(((((...(((.....)))...)))))..)))))))........-.. ( -26.80) >DroSec_CAF1 13925 119 - 1 AGUUGCCAGGCUUCUUUUUGCAACACAGAAAAUGCAAUAUAGAUCGUAAUCUUAAUUGCAUUUAGAGUGCCCCGCAUUGUAGGUUAAUAAUCAUAAAUGUUAUGGUAAUAGUUACGA-GU .((((((((((((((.((((.....))))(((((((((..((((....))))..))))))))))))).)))..(((((...(((.....)))...)))))..)))))))........-.. ( -26.80) >DroSim_CAF1 22898 119 - 1 AGUAGCCAGGCUUCUUUUUGCAACACAGAAAAUGCAAUAUAGAUCGUAAUCUUAAUUGCAUUUAGAGUGCCCCGCAUUGUAGGUUAAUAAUCAUAAAUGUUAUGGUAAUAGUUACGA-GU ....((..(((((((.((((.....))))(((((((((..((((....))))..))))))))))))).)))..)).((((((.(.....((((((.....))))))...).))))))-.. ( -25.40) >DroEre_CAF1 16526 119 - 1 AGUGGCCAGGCUUCUUUUUGCAACACAGAAAAUGCAAUAUAGAUCGUAAUCUUAAUUGCAUUUAGAGUGCCCCGCAUUGUGGGUUAAUAAUCAUAAAUGUUAUGGUAAUAGUUACGA-GU .((((...(((((((.((((.....))))(((((((((..((((....))))..))))))))))))).))))))).((((((.(.....((((((.....))))))...).))))))-.. ( -27.00) >DroYak_CAF1 20719 119 - 1 AGUUGCCAGGCUCCUUUUUGCAACACAGAAAAUGCAAUAUAGAUCGUAAUCUUAAUUGCAUUUAGAGUGCCCCGCAUUGUAGGUUAAUAAUCAUAAAUGUUAUGGUAAUAGUUACGA-GU .((((((((((.(((.((((.....))))(((((((((..((((....))))..))))))))))).).)))..(((((...(((.....)))...)))))..)))))))........-.. ( -25.30) >DroAna_CAF1 49715 120 - 1 AGUUGCCAGGUUUCUUUUUGCAACACAGAAAAUGUAAUAUAGAUCGUAAUCUUAAUUGCAUUUAGAGUGCCCCGCAUUGUGGGUUAAUAAUCAUAAAUGCUACGGUAAUAGUUACGGGGC .(((((.(((.....))).))))).....(((((((((..((((....))))..))))))))).....((((((..((((((........)))))).(((....))).......)))))) ( -31.90) >consensus AGUUGCCAGGCUUCUUUUUGCAACACAGAAAAUGCAAUAUAGAUCGUAAUCUUAAUUGCAUUUAGAGUGCCCCGCAUUGUAGGUUAAUAAUCAUAAAUGUUAUGGUAAUAGUUACGA_GU .((((((((((((((.((((.....))))(((((((((..((((....))))..))))))))))))).)))..(((((...(((.....)))...)))))..)))))))........... (-24.92 = -25.17 + 0.25)


| Location | 13,606,779 – 13,606,899 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.44 |
| Mean single sequence MFE | -28.33 |
| Consensus MFE | -25.77 |
| Energy contribution | -25.72 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.91 |
| SVM decision value | 3.16 |
| SVM RNA-class probability | 0.998601 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13606779 120 + 23771897 ACAAUGCGGGGCACUCUAAAUGCAAUUAAGAUUACGAUCUAUAUUGCAUUUUCUGUGUUGCAAAAAGAAGCCUGGCAACUCUGCCACGACAAACAAUUAAAAACAAUGCGCCUCCAUCUU ...(((.(((((((...(((((((((..((((....))))..)))))))))...))(((((........)).(((((....))))).)))...................))))))))... ( -28.90) >DroSec_CAF1 13964 120 + 1 ACAAUGCGGGGCACUCUAAAUGCAAUUAAGAUUACGAUCUAUAUUGCAUUUUCUGUGUUGCAAAAAGAAGCCUGGCAACUCUGCCACGACAAACAAUUAAAAACAAUGCGCCUCCAUCUU ...(((.(((((((...(((((((((..((((....))))..)))))))))...))(((((........)).(((((....))))).)))...................))))))))... ( -28.90) >DroSim_CAF1 22937 120 + 1 ACAAUGCGGGGCACUCUAAAUGCAAUUAAGAUUACGAUCUAUAUUGCAUUUUCUGUGUUGCAAAAAGAAGCCUGGCUACUCUGCCACGACAAACAAUUAAAAACAAUGCGCCUCCAUCUU ...(((.(((((((...(((((((((..((((....))))..)))))))))...))(((((........)).((((......)))).)))...................))))))))... ( -28.50) >DroEre_CAF1 16565 120 + 1 ACAAUGCGGGGCACUCUAAAUGCAAUUAAGAUUACGAUCUAUAUUGCAUUUUCUGUGUUGCAAAAAGAAGCCUGGCCACUCUGCCACGACAAACAAUUAAAAACAAUGCGCCUCCAUCUU ...(((.(((((((...(((((((((..((((....))))..)))))))))...))(((((........)).((((......)))).)))...................))))))))... ( -27.30) >DroYak_CAF1 20758 120 + 1 ACAAUGCGGGGCACUCUAAAUGCAAUUAAGAUUACGAUCUAUAUUGCAUUUUCUGUGUUGCAAAAAGGAGCCUGGCAACUCUGCCACGACAAACAAUUAAAAACAAUGCGCCUCCAUCUU ...(((.(((((((...(((((((((..((((....))))..)))))))))...))...(((...((....))((((....)))).....................)))))))))))... ( -30.30) >DroAna_CAF1 49755 120 + 1 ACAAUGCGGGGCACUCUAAAUGCAAUUAAGAUUACGAUCUAUAUUACAUUUUCUGUGUUGCAAAAAGAAACCUGGCAACUCUGCUACGACAAACAAUUAAAAACAAUGCACUCCCGCAGU ....((((((((((...(((((.(((..((((....))))..))).)))))...))))((((...((....))((((....)))).....................))))..)))))).. ( -26.10) >consensus ACAAUGCGGGGCACUCUAAAUGCAAUUAAGAUUACGAUCUAUAUUGCAUUUUCUGUGUUGCAAAAAGAAGCCUGGCAACUCUGCCACGACAAACAAUUAAAAACAAUGCGCCUCCAUCUU ...(((.(((((((...(((((((((..((((....))))..)))))))))...))...(((...((....))(((......))).....................)))))))))))... (-25.77 = -25.72 + -0.05)



| Location | 13,606,779 – 13,606,899 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.44 |
| Mean single sequence MFE | -33.45 |
| Consensus MFE | -30.44 |
| Energy contribution | -30.05 |
| Covariance contribution | -0.39 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.81 |
| SVM RNA-class probability | 0.978254 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13606779 120 - 23771897 AAGAUGGAGGCGCAUUGUUUUUAAUUGUUUGUCGUGGCAGAGUUGCCAGGCUUCUUUUUGCAACACAGAAAAUGCAAUAUAGAUCGUAAUCUUAAUUGCAUUUAGAGUGCCCCGCAUUGU ..(.(((.(((((..((((...((..(...(((.((((......)))))))..)..))...))))....(((((((((..((((....))))..)))))))))...)))))))))..... ( -33.10) >DroSec_CAF1 13964 120 - 1 AAGAUGGAGGCGCAUUGUUUUUAAUUGUUUGUCGUGGCAGAGUUGCCAGGCUUCUUUUUGCAACACAGAAAAUGCAAUAUAGAUCGUAAUCUUAAUUGCAUUUAGAGUGCCCCGCAUUGU ..(.(((.(((((..((((...((..(...(((.((((......)))))))..)..))...))))....(((((((((..((((....))))..)))))))))...)))))))))..... ( -33.10) >DroSim_CAF1 22937 120 - 1 AAGAUGGAGGCGCAUUGUUUUUAAUUGUUUGUCGUGGCAGAGUAGCCAGGCUUCUUUUUGCAACACAGAAAAUGCAAUAUAGAUCGUAAUCUUAAUUGCAUUUAGAGUGCCCCGCAUUGU ..(.(((.(((((..............(((((.((.((((((.(((...)))...)))))).)))))))(((((((((..((((....))))..)))))))))...)))))))))..... ( -34.20) >DroEre_CAF1 16565 120 - 1 AAGAUGGAGGCGCAUUGUUUUUAAUUGUUUGUCGUGGCAGAGUGGCCAGGCUUCUUUUUGCAACACAGAAAAUGCAAUAUAGAUCGUAAUCUUAAUUGCAUUUAGAGUGCCCCGCAUUGU ..(.(((.(((((..............(((((.((.((((((.(((...)))...)))))).)))))))(((((((((..((((....))))..)))))))))...)))))))))..... ( -34.20) >DroYak_CAF1 20758 120 - 1 AAGAUGGAGGCGCAUUGUUUUUAAUUGUUUGUCGUGGCAGAGUUGCCAGGCUCCUUUUUGCAACACAGAAAAUGCAAUAUAGAUCGUAAUCUUAAUUGCAUUUAGAGUGCCCCGCAUUGU ..(.(((.(((((.((((.........(((((....)))))(((((.((....))....))))))))).(((((((((..((((....))))..)))))))))...)))))))))..... ( -32.80) >DroAna_CAF1 49755 120 - 1 ACUGCGGGAGUGCAUUGUUUUUAAUUGUUUGUCGUAGCAGAGUUGCCAGGUUUCUUUUUGCAACACAGAAAAUGUAAUAUAGAUCGUAAUCUUAAUUGCAUUUAGAGUGCCCCGCAUUGU ..((((((.(..(.((((......(((((......))))).(((((.(((.....))).))))))))).(((((((((..((((....))))..)))))))))...)..))))))).... ( -33.30) >consensus AAGAUGGAGGCGCAUUGUUUUUAAUUGUUUGUCGUGGCAGAGUUGCCAGGCUUCUUUUUGCAACACAGAAAAUGCAAUAUAGAUCGUAAUCUUAAUUGCAUUUAGAGUGCCCCGCAUUGU ....(((.(((((..............(((((.((.((((((..((...))....)))))).)))))))(((((((((..((((....))))..)))))))))...))))))))...... (-30.44 = -30.05 + -0.39)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:35:41 2006