
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,602,872 – 13,603,024 |
| Length | 152 |
| Max. P | 0.987162 |

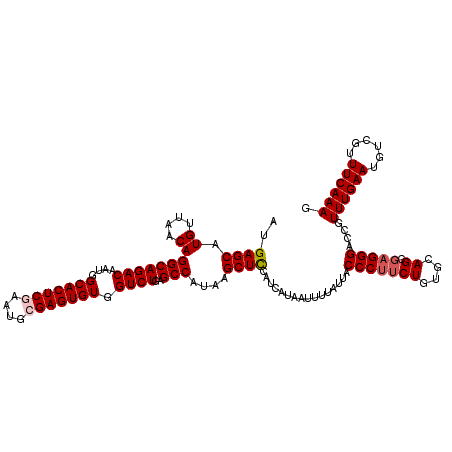
| Location | 13,602,872 – 13,602,992 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.61 |
| Mean single sequence MFE | -34.45 |
| Consensus MFE | -30.75 |
| Energy contribution | -31.28 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.15 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.535122 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13602872 120 - 23771897 AUGAGCAUGUUAACAGGCAGACAAUGGCACUCAAAUGCGAGUGUGGUCUGAGCCAUAAGCUCCAUCAUAAUUUUAUUACCCUUCUGUGCAGCGAGGGACCGUUUGAAUGUCGUUUCAAAG ....((.((....)).)).((.(((((((.(((((((.(..((((((..((((.....)))).)))))).........(((((((....)).))))).)))))))).))))))))).... ( -32.90) >DroSec_CAF1 9791 120 - 1 AUGAGCAUGUUAACAGGCAGACAAUGGCACUCGAAUGCGAGUGUGGUCUGAGCCAUAAGCUCCAUCAUAAUUUUAUUACCCUUCUGUGCAGCGAGGGACCGUUUGAAUGUCGUUUCAAAG .((((.(((...(((..(((((....(((((((....)))))))((((.((((.....)))).................((((((....)).)))))))))))))..))))))))))... ( -34.70) >DroSim_CAF1 19033 120 - 1 AUGAGCAUGUUAACAGGCAGACAAUGGCACUCGAAUGCGAGUGUGGUCUGAGCCAUAAGCUCCAUCAUAAUUUUAUUACCCUUCUGUGCAGCGAGGGACCGUUUGAAUGUCGUUUCAAAG .((((.(((...(((..(((((....(((((((....)))))))((((.((((.....)))).................((((((....)).)))))))))))))..))))))))))... ( -34.70) >DroEre_CAF1 12606 120 - 1 AUGAGCAUGUUAACAGGCAGACAAUGGCACUCGAAUGCGAGUGUGGUCUGAGCCAUAAGCUCCAUCAUAAUUUUAUUACCCUUCUGUGCAGCGAGGGACCGUUUGAAUGUCGUUUCAAAG .((((.(((...(((..(((((....(((((((....)))))))((((.((((.....)))).................((((((....)).)))))))))))))..))))))))))... ( -34.70) >DroYak_CAF1 16031 120 - 1 AUGAGCAUGUUAACAGGCAGACAAUGGCACUCGAAUGCGAGUGUGGUCUGAGCCAUAAGCUCCAUCAUAAUUUUAUUACCCUUCUGUGCAGCGAGGGACCGUUUGAAUGUCGUUUCAAAG .((((.(((...(((..(((((....(((((((....)))))))((((.((((.....)))).................((((((....)).)))))))))))))..))))))))))... ( -34.70) >DroAna_CAF1 45741 119 - 1 AUGAGCAUGUUAACAGGCAGACAAUGGCACUCAAAUGCGAGUGUGGUCUGAGCCAUAAGCUUCAUCAUAAUUUUAUUACCCCGCUCUGCAGCGAGGGAGC-UUUGAAUGUCCUUUCAAAG ((((((.((....))(((((((....((((((......)))))).))))..)))....)).)))).............(((((((....)))).)))..(-((((((......))))))) ( -35.00) >consensus AUGAGCAUGUUAACAGGCAGACAAUGGCACUCGAAUGCGAGUGUGGUCUGAGCCAUAAGCUCCAUCAUAAUUUUAUUACCCUUCUGUGCAGCGAGGGACCGUUUGAAUGUCGUUUCAAAG ..((((.((....))(((((((....(((((((....))))))).))))..)))....))))................(((((((....)).)))))....((((((......)))))). (-30.75 = -31.28 + 0.53)


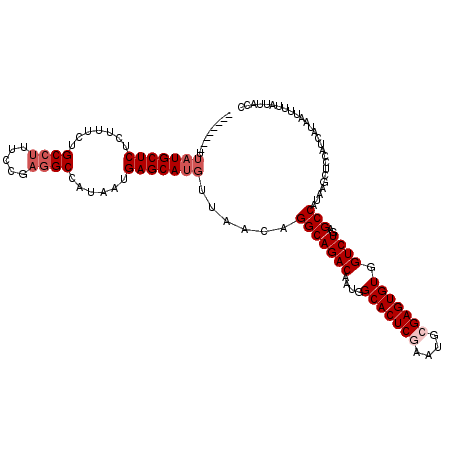
| Location | 13,602,912 – 13,603,024 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.59 |
| Mean single sequence MFE | -31.15 |
| Consensus MFE | -28.09 |
| Energy contribution | -28.54 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.818958 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13602912 112 + 23771897 GGUAAUAAAAUUAUGAUGGAGCUUAUGGCUCAGACCACACUCGCAUUUGAGUGCCAUUGUCUGCCUGUUAACAUGCUCAUUAUGGCCUCGGAAAGGCAGAAAGAGAGCAUAA-------- (((...............((((.....))))((((..((((((....)))))).....))))))).......((((((......((((.....)))).......))))))..-------- ( -29.72) >DroSec_CAF1 9831 112 + 1 GGUAAUAAAAUUAUGAUGGAGCUUAUGGCUCAGACCACACUCGCAUUCGAGUGCCAUUGUCUGCCUGUUAACAUGCUCAUUAUGGCCUCGGAAAGGCAGAAAGAGAGCAUAA-------- (((...............((((.....))))((((..((((((....)))))).....))))))).......((((((......((((.....)))).......))))))..-------- ( -31.02) >DroSim_CAF1 19073 112 + 1 GGUAAUAAAAUUAUGAUGGAGCUUAUGGCUCAGACCACACUCGCAUUCGAGUGCCAUUGUCUGCCUGUUAACAUGCUCAUUAUGGCCUCGGAAAGGCAGAAAGAGAGCAUAA-------- (((...............((((.....))))((((..((((((....)))))).....))))))).......((((((......((((.....)))).......))))))..-------- ( -31.02) >DroEre_CAF1 12646 112 + 1 GGUAAUAAAAUUAUGAUGGAGCUUAUGGCUCAGACCACACUCGCAUUCGAGUGCCAUUGUCUGCCUGUUAACAUGCUCAUUAUGGCCUCGGAAAGGCAGAAAGAGAGCAUAA-------- (((...............((((.....))))((((..((((((....)))))).....))))))).......((((((......((((.....)))).......))))))..-------- ( -31.02) >DroYak_CAF1 16071 112 + 1 GGUAAUAAAAUUAUGAUGGAGCUUAUGGCUCAGACCACACUCGCAUUCGAGUGCCAUUGUCUGCCUGUUAACAUGCUCAUUAUGGCCUCGGAAAGGCAGAAAGAGAGCAUAA-------- (((...............((((.....))))((((..((((((....)))))).....))))))).......((((((......((((.....)))).......))))))..-------- ( -31.02) >DroAna_CAF1 45780 116 + 1 GGUAAUAAAAUUAUGAUGAAGCUUAUGGCUCAGACCACACUCGCAUUUGAGUGCCAUUGUCUGCCUGUUAACAUGCUCAUUAUGGC----GAAAGGCAGAAACAGAGCAAUGGGCCAGAC ....................((((((.((((......((((((....))))))......(((((((.((..((((.....))))..----)).)))))))....)))).))))))..... ( -33.10) >consensus GGUAAUAAAAUUAUGAUGGAGCUUAUGGCUCAGACCACACUCGCAUUCGAGUGCCAUUGUCUGCCUGUUAACAUGCUCAUUAUGGCCUCGGAAAGGCAGAAAGAGAGCAUAA________ (((...............((((.....))))((((..((((((....)))))).....))))))).......((((((......((((.....)))).......)))))).......... (-28.09 = -28.54 + 0.44)

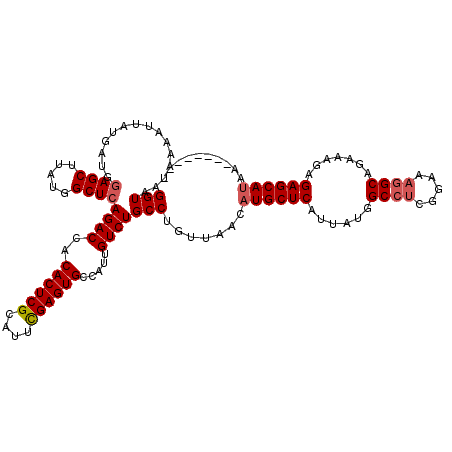
| Location | 13,602,912 – 13,603,024 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.59 |
| Mean single sequence MFE | -31.37 |
| Consensus MFE | -28.32 |
| Energy contribution | -29.32 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.90 |
| SVM decision value | 2.07 |
| SVM RNA-class probability | 0.987162 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13602912 112 - 23771897 --------UUAUGCUCUCUUUCUGCCUUUCCGAGGCCAUAAUGAGCAUGUUAACAGGCAGACAAUGGCACUCAAAUGCGAGUGUGGUCUGAGCCAUAAGCUCCAUCAUAAUUUUAUUACC --------.(((((((.......(((((...)))))......)))))))......(((((((....((((((......)))))).)))))(((.....)))))................. ( -30.32) >DroSec_CAF1 9831 112 - 1 --------UUAUGCUCUCUUUCUGCCUUUCCGAGGCCAUAAUGAGCAUGUUAACAGGCAGACAAUGGCACUCGAAUGCGAGUGUGGUCUGAGCCAUAAGCUCCAUCAUAAUUUUAUUACC --------.(((((((.......(((((...)))))......)))))))......(((((((....(((((((....))))))).)))))(((.....)))))................. ( -32.32) >DroSim_CAF1 19073 112 - 1 --------UUAUGCUCUCUUUCUGCCUUUCCGAGGCCAUAAUGAGCAUGUUAACAGGCAGACAAUGGCACUCGAAUGCGAGUGUGGUCUGAGCCAUAAGCUCCAUCAUAAUUUUAUUACC --------.(((((((.......(((((...)))))......)))))))......(((((((....(((((((....))))))).)))))(((.....)))))................. ( -32.32) >DroEre_CAF1 12646 112 - 1 --------UUAUGCUCUCUUUCUGCCUUUCCGAGGCCAUAAUGAGCAUGUUAACAGGCAGACAAUGGCACUCGAAUGCGAGUGUGGUCUGAGCCAUAAGCUCCAUCAUAAUUUUAUUACC --------.(((((((.......(((((...)))))......)))))))......(((((((....(((((((....))))))).)))))(((.....)))))................. ( -32.32) >DroYak_CAF1 16071 112 - 1 --------UUAUGCUCUCUUUCUGCCUUUCCGAGGCCAUAAUGAGCAUGUUAACAGGCAGACAAUGGCACUCGAAUGCGAGUGUGGUCUGAGCCAUAAGCUCCAUCAUAAUUUUAUUACC --------.(((((((.......(((((...)))))......)))))))......(((((((....(((((((....))))))).)))))(((.....)))))................. ( -32.32) >DroAna_CAF1 45780 116 - 1 GUCUGGCCCAUUGCUCUGUUUCUGCCUUUC----GCCAUAAUGAGCAUGUUAACAGGCAGACAAUGGCACUCAAAUGCGAGUGUGGUCUGAGCCAUAAGCUUCAUCAUAAUUUUAUUACC (((((......(((((.(((...((.....----))...))))))))......)))))((((....((((((......)))))).))))((((.....)))).................. ( -28.60) >consensus ________UUAUGCUCUCUUUCUGCCUUUCCGAGGCCAUAAUGAGCAUGUUAACAGGCAGACAAUGGCACUCGAAUGCGAGUGUGGUCUGAGCCAUAAGCUCCAUCAUAAUUUUAUUACC .........(((((((.......((((.....))))......)))))))......(((((((....(((((((....))))))).))))..))).......................... (-28.32 = -29.32 + 1.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:35:36 2006