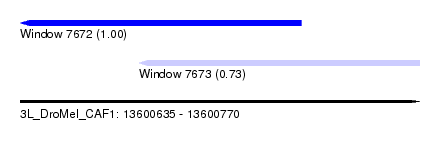
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,600,635 – 13,600,770 |
| Length | 135 |
| Max. P | 0.997038 |

| Location | 13,600,635 – 13,600,730 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 85.77 |
| Mean single sequence MFE | -30.02 |
| Consensus MFE | -26.54 |
| Energy contribution | -26.82 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.58 |
| Structure conservation index | 0.88 |
| SVM decision value | 2.79 |
| SVM RNA-class probability | 0.997038 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13600635 95 - 23771897 AUGUUGUAUAACAGCAGCGGCAACAAGCCGUCCUGCUG---GUGGGCGUGGCACACACGCACUCACACUCAUACGCGCACAAUAACACAUCGACAUAC ((((((.....((((((((((.....))))..))))))---(((((((((.....))))).).)))........................)))))).. ( -32.10) >DroSec_CAF1 7446 94 - 1 AUGUUGUAUAACAGCAGCGGAAACAAGCCGUCCUGCU----GUGGGCGUGGCACAAUCGCACUCACACUCAUACGCGCACAAUAACACAUCGACAUAC ((((((......((((((((.......)))..)))))----(((.(((((((......))...........))))).)))..........)))))).. ( -23.60) >DroSim_CAF1 16807 98 - 1 AUGUUGUAUAACAGCAGCGGCAACAAGCCGUCCUGCUGCCUGUGGGCGUGGCCCACUCGCACUCACACUCAUACGCGCACAAUAACACAUCGACACAC .((((((....((((((((((.....))))..))))))..((((.(((((.....................))))).))))))))))........... ( -29.80) >DroEre_CAF1 10336 89 - 1 AUGUUGCAUAACAGCAGCGGCAACAAGCCGUCCUGCUG---GUGGGCGUGGCAGAC------GCACACGCACACGCACACAAUCACACUUCGACAUAC .((((((....((((((((((.....))))..))))))---(((.(((((((....------)).))))).)))))).)))................. ( -35.20) >DroYak_CAF1 13692 83 - 1 AUGUUGUAUAACAGCGGCGGCAACAAGCCGGCCUGCUG---GUGGGCGUGGC------------ACACUCAUACGCACACAAUAACACAUCGACAUGC ((((((.....((((((((((.....))).))).))))---(((.(((((..------------.......))))).)))..........)))))).. ( -29.40) >consensus AUGUUGUAUAACAGCAGCGGCAACAAGCCGUCCUGCUG___GUGGGCGUGGCACAC_CGCACUCACACUCAUACGCGCACAAUAACACAUCGACAUAC ((((((.....((((((((((.....))))..))))))...(((.(((((.....................))))).)))..........)))))).. (-26.54 = -26.82 + 0.28)

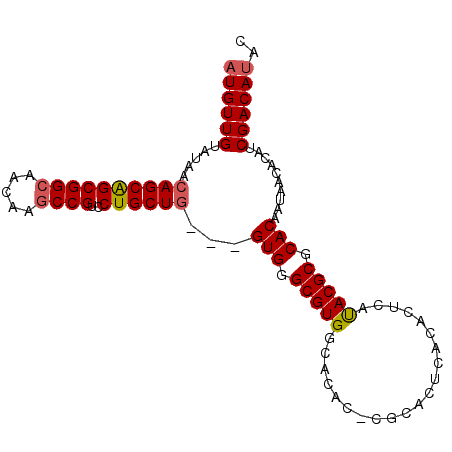
| Location | 13,600,675 – 13,600,770 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.81 |
| Mean single sequence MFE | -28.87 |
| Consensus MFE | -22.91 |
| Energy contribution | -23.60 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.730999 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
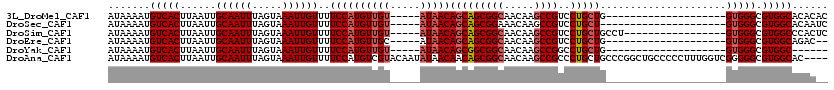
>3L_DroMel_CAF1 13600675 95 - 23771897 AUAAAAUGUCACUUAAUUGCAAUUUAGUAAAUUGUUUUCCAUGUUGU-----AUAACAGCAGCGGCAACAAGCCGUCCUGCUG--------------------GUGGGCGUGGCACACAC ......((((((......((((((.....))))))..........((-----....((((((((((.....))))..))))))--------------------....))))))))..... ( -27.00) >DroSec_CAF1 7486 94 - 1 AUAAAAUGUCACUUAAUUGCAAUUUAGUAAAUUGUUUUCCAUGUUGU-----AUAACAGCAGCGGAAACAAGCCGUCCUGCU---------------------GUGGGCGUGGCACAAUC ......((((((....((((......)))).((((((((..(((((.-----....)))))..))))))))...((((....---------------------..))))))))))..... ( -26.20) >DroSim_CAF1 16847 98 - 1 AUAAAAUGUCACUUAAUUGCAAUUUAGUAAAUUGUUUUCCAUGUUGU-----AUAACAGCAGCGGCAACAAGCCGUCCUGCUGCCU-----------------GUGGGCGUGGCCCACUC .....(((..((......((((((.....)))))).......))..)-----))..((((((((((.....))))..))))))...-----------------((((((...)))))).. ( -27.82) >DroEre_CAF1 10372 93 - 1 AUAAAAUGUCACUUAAUUGCAAUUUAGUAAAUUGUUUUCCAUGUUGC-----AUAACAGCAGCGGCAACAAGCCGUCCUGCUG--------------------GUGGGCGUGGCAGAC-- ......((((((......((((((.....))))))..........((-----....((((((((((.....))))..))))))--------------------....))))))))...-- ( -28.90) >DroYak_CAF1 13726 89 - 1 AUAAAAUGUCACUUAAUUGCAAUUUAGUAAAUUGUUUUCCAUGUUGU-----AUAACAGCGGCGGCAACAAGCCGGCCUGCUG--------------------GUGGGCGUGGC------ .......(((((....((((......)))).(((((..((.(((((.-----....)))))..)).)))))(((.(((....)--------------------)).))))))))------ ( -26.20) >DroAna_CAF1 43324 116 - 1 AUAAAAUGUCACUUAAUUGCAAUUUAGUAAAUUGUUUUCCAUGUCGUACAAUAUAACAACAGCGGCAACAAGCCGCCCUGCUGCCCGGCUGCCCCCUUUGGUCGGGGGCGUGGCAC---- ......((((((......((....(((((..(((((.....((.....))....)))))..(((((.....)))))..)))))....)).((((((.......)))))))))))).---- ( -37.10) >consensus AUAAAAUGUCACUUAAUUGCAAUUUAGUAAAUUGUUUUCCAUGUUGU_____AUAACAGCAGCGGCAACAAGCCGUCCUGCUG____________________GUGGGCGUGGCACAC__ .......(((((......((((((.....))))))..((((((((((.....)))))(((((((((.....))))..))))).....................))))).)))))...... (-22.91 = -23.60 + 0.69)
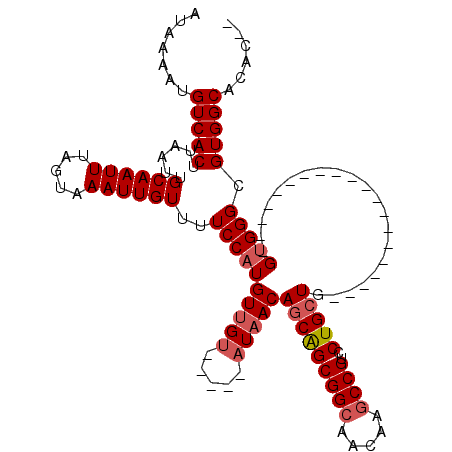
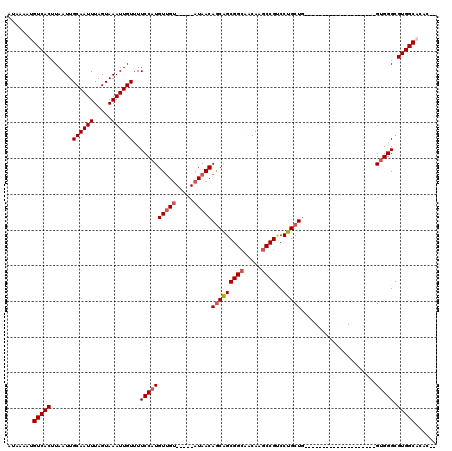
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:35:29 2006