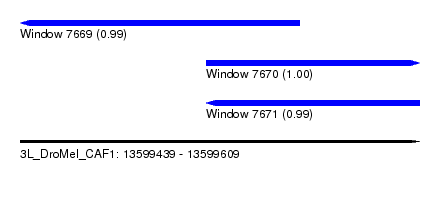
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,599,439 – 13,599,609 |
| Length | 170 |
| Max. P | 0.999737 |

| Location | 13,599,439 – 13,599,558 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.02 |
| Mean single sequence MFE | -34.53 |
| Consensus MFE | -28.83 |
| Energy contribution | -29.52 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.15 |
| Mean z-score | -3.71 |
| Structure conservation index | 0.83 |
| SVM decision value | 2.23 |
| SVM RNA-class probability | 0.990819 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13599439 119 - 23771897 AACCCAUAUAUUCAUAAUUAAUUGUUUGCCUGAGCAGACUUCUAAGUGAAUUUUCGUUCCCUUAUUAAUGGCCCACUACCUCGGGCCGUCUGCUUUGGCCCG-AAUGGCUAAUUAAACAU ..............((((((.(..((((((.((((((((...((((.((((....)))).)))).....(((((........))))))))))))).)))..)-))..).))))))..... ( -38.00) >DroSec_CAF1 6277 120 - 1 AACCCAUAUAUUCAUAAUUAAUUGUUUGCCUGAGCAGACUUCUAAGUGAAUUUUCGUUCCCUUAUUAAUGGCCCACUACCUCGGGCCGUCUGCUUUGGCCCGGAAUGGCUAAUUAAACAU ..............((((((.(..((((((.((((((((...((((.((((....)))).)))).....(((((........))))))))))))).)))...)))..).))))))..... ( -37.60) >DroSim_CAF1 15632 120 - 1 AACCCAUAUAUUCAUAAUUAAUUGUUUGCCUGAGCAGACUUCUAAGUGAAUUUUCGUUCCCUUAUUAAUGGCCCACUACCUCGGGCCGUCUGCUUUGGCCCGGAAUGGCUAAUUAAACAU ..............((((((.(..((((((.((((((((...((((.((((....)))).)))).....(((((........))))))))))))).)))...)))..).))))))..... ( -37.60) >DroEre_CAF1 7489 106 - 1 AACCCACAUAUUCAUAAUUAAUUGUUUGCCUGAGCAGACUUCUAAGUGAAUUUGCGUUCCCUUAUUAAUG-------------GGCCGUCUGUUCUGGCCCG-AAUGGCUAAUUAAACAU ..............((((((.(..((((((.(((((((((((.....)))........(((........)-------------))..)))))))).)))..)-))..).))))))..... ( -26.70) >DroYak_CAF1 12448 119 - 1 AACCCACAUAUUCAUAAUUAAUUGUUUGCCCGAGCAGACUUCUAAGUGAAUUUUCGUUCCCUUAUUAAUGGCCCACUACCUCGGGCCGUCUGCUUUGGCCCG-AAUGGCUAAUUAAACAU ..............((((((.(..((((((.((((((((...((((.((((....)))).)))).....(((((........))))))))))))).)))..)-))..).))))))..... ( -38.00) >DroAna_CAF1 42078 117 - 1 GGCCCACAUAUCCAUUAUUAAUUGUCUUCCUGGGCAGACUUCUAAGUGAAUU-UCGUUCCCUUAUUAAUGACAGGCCGCCUCGGGCUGUCUGUUCUGGUCUG-AGUGGCUAAUUAAACA- ((((.((...((((................))))((((((..((((.((((.-..)))).)))).....(((((.(((...))).)))))......))))))-.)))))).........- ( -29.29) >consensus AACCCACAUAUUCAUAAUUAAUUGUUUGCCUGAGCAGACUUCUAAGUGAAUUUUCGUUCCCUUAUUAAUGGCCCACUACCUCGGGCCGUCUGCUUUGGCCCG_AAUGGCUAAUUAAACAU ..............((((((.(..((.(((.((((((((...((((.((((....)))).)))).....(((((........))))))))))))).)))....))..).))))))..... (-28.83 = -29.52 + 0.68)


| Location | 13,599,518 – 13,599,609 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.74 |
| Mean single sequence MFE | -25.28 |
| Consensus MFE | -24.24 |
| Energy contribution | -23.68 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.76 |
| Structure conservation index | 0.96 |
| SVM decision value | 3.98 |
| SVM RNA-class probability | 0.999737 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13599518 91 + 23771897 AGUCUGCUCAGGCAAACAAUUAAUUAUGAAUAUAUGGGUUAAUGGAAAACCCAGACUGAUUUUCCAGACAAAGUGCUAUUCAGCGCUUUAA----------------------------- .(((((.((((.....((........))......((((((.......))))))..)))).....)))))((((((((....))))))))..----------------------------- ( -23.70) >DroSec_CAF1 6357 91 + 1 AGUCUGCUCAGGCAAACAAUUAAUUAUGAAUAUAUGGGUUAAUGGAAAACCCAGACUGAUUUUCCAGACAAAGUGCUAUUCAGCGCUUUAA----------------------------- .(((((.((((.....((........))......((((((.......))))))..)))).....)))))((((((((....))))))))..----------------------------- ( -23.70) >DroSim_CAF1 15712 91 + 1 AGUCUGCUCAGGCAAACAAUUAAUUAUGAAUAUAUGGGUUAAUGGAAAACCCAGACUGAUUUUCCAGACAAAGUGCUAUUCAGCGCUUUAA----------------------------- .(((((.((((.....((........))......((((((.......))))))..)))).....)))))((((((((....))))))))..----------------------------- ( -23.70) >DroEre_CAF1 7555 120 + 1 AGUCUGCUCAGGCAAACAAUUAAUUAUGAAUAUGUGGGUUAAUGGAAAGCCCAGACUGAUUUUCCAGACAAAGUGCUAUUCAGCACUUUAAUAUUUUUGACGGAAAAUCGGGAAAACGAC .((.(((....))).))...............(.((((((.......)))))).)((((((((((....((((((((....))))))))..((....))..))))))))))......... ( -31.80) >DroYak_CAF1 12527 91 + 1 AGUCUGCUCGGGCAAACAAUUAAUUAUGAAUAUGUGGGUUAAUGGAAAACCCAGACUGAUUUUCCAGACAAAGUGCUAUUCAGCACUUUAA----------------------------- .(((((.((((.....((........))....(.((((((.......)))))).))))).....)))))((((((((....))))))))..----------------------------- ( -23.50) >consensus AGUCUGCUCAGGCAAACAAUUAAUUAUGAAUAUAUGGGUUAAUGGAAAACCCAGACUGAUUUUCCAGACAAAGUGCUAUUCAGCGCUUUAA_____________________________ .(((((.((((.....((........))......((((((.......))))))..)))).....)))))((((((((....))))))))............................... (-24.24 = -23.68 + -0.56)

| Location | 13,599,518 – 13,599,609 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.74 |
| Mean single sequence MFE | -23.85 |
| Consensus MFE | -20.52 |
| Energy contribution | -20.72 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.86 |
| SVM decision value | 2.33 |
| SVM RNA-class probability | 0.992401 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13599518 91 - 23771897 -----------------------------UUAAAGCGCUGAAUAGCACUUUGUCUGGAAAAUCAGUCUGGGUUUUCCAUUAACCCAUAUAUUCAUAAUUAAUUGUUUGCCUGAGCAGACU -----------------------------.(((((.(((....))).)))))..(((((((((......))))))))).........................((((((....)))))). ( -21.50) >DroSec_CAF1 6357 91 - 1 -----------------------------UUAAAGCGCUGAAUAGCACUUUGUCUGGAAAAUCAGUCUGGGUUUUCCAUUAACCCAUAUAUUCAUAAUUAAUUGUUUGCCUGAGCAGACU -----------------------------.(((((.(((....))).)))))..(((((((((......))))))))).........................((((((....)))))). ( -21.50) >DroSim_CAF1 15712 91 - 1 -----------------------------UUAAAGCGCUGAAUAGCACUUUGUCUGGAAAAUCAGUCUGGGUUUUCCAUUAACCCAUAUAUUCAUAAUUAAUUGUUUGCCUGAGCAGACU -----------------------------.(((((.(((....))).)))))..(((((((((......))))))))).........................((((((....)))))). ( -21.50) >DroEre_CAF1 7555 120 - 1 GUCGUUUUCCCGAUUUUCCGUCAAAAAUAUUAAAGUGCUGAAUAGCACUUUGUCUGGAAAAUCAGUCUGGGCUUUCCAUUAACCCACAUAUUCAUAAUUAAUUGUUUGCCUGAGCAGACU (((........(((((((((..........(((((((((....)))))))))..))))))))).((..((((...(.(((((...............))))).)...))))..)).))). ( -29.36) >DroYak_CAF1 12527 91 - 1 -----------------------------UUAAAGUGCUGAAUAGCACUUUGUCUGGAAAAUCAGUCUGGGUUUUCCAUUAACCCACAUAUUCAUAAUUAAUUGUUUGCCCGAGCAGACU -----------------------------.(((((((((....)))))))))..(((((((((......))))))))).........................((((((....)))))). ( -25.40) >consensus _____________________________UUAAAGCGCUGAAUAGCACUUUGUCUGGAAAAUCAGUCUGGGUUUUCCAUUAACCCAUAUAUUCAUAAUUAAUUGUUUGCCUGAGCAGACU ..............................(((((.(((....))).)))))..(((((((((......))))))))).........................((((((....)))))). (-20.52 = -20.72 + 0.20)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:35:27 2006