
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,598,283 – 13,598,414 |
| Length | 131 |
| Max. P | 0.756000 |

| Location | 13,598,283 – 13,598,384 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 96.09 |
| Mean single sequence MFE | -25.06 |
| Consensus MFE | -23.50 |
| Energy contribution | -23.70 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.11 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.584481 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
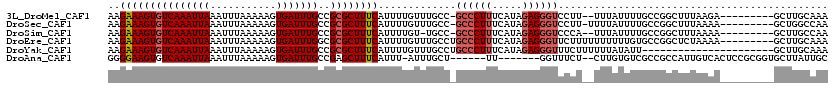
>3L_DroMel_CAF1 13598283 101 + 23771897 UGAUGCUCGCAGCUCAGAGAGAAAAGAAAGUGUCAAAUUAAAUUUAAAAAGUGAUUUGCCGCGCUUUCAUUUUGUUUGCC-GCCCUUUCAUAGAGGGUCCUU- (((.((.....))))).(((.(((((((((((((((((((...........)))))))..)))))))).)))).)))...-((((((.....))))))....- ( -24.30) >DroSec_CAF1 5139 101 + 1 UGAUGCUCGCAGCUCAGAGAGAAAAGAAAGUGUCAAAUUAAAUUUAAAAAGUGAUUUGCCGCGCUUUCAUUUUGUUUGCC-GCCCUUUCAUAGAGGGUCCUU- (((.((.....))))).(((.(((((((((((((((((((...........)))))))..)))))))).)))).)))...-((((((.....))))))....- ( -24.30) >DroSim_CAF1 14517 100 + 1 UGAUGCUCGCAGCUCAGAGAGAAAAGAAAGUGUCAAAUUAAAUUUAAAAAGUGAUUUGCCGCGCUUUCAUUUUGU-UGCC-GCCCUUUCAUAGAGGGUCCCA- ........(((((((.....))...(((((((((((((((...........)))))))..)))))))).....))-))).-((((((.....))))))....- ( -24.70) >DroEre_CAF1 6340 103 + 1 UGAUGCUCGCAGCUCAGAGAGAAAAGAAAGUGUCAAAUUAAAUUUAAAAAGUGAUUUGGCGCGCUUUCAUUUUGUUUGCCUGCCCUUUCAUAGAGGGUUCUUU ........((((..(((((.((((.(...(((((((((((...........))))))))))).))))).))))).))))..((((((.....))))))..... ( -26.80) >DroYak_CAF1 11307 103 + 1 UGAUGCUCGCAGCUCAGAGAGAAAAGAAAGUGUCAAAUUAAAUUUAAAAAGUGAUUUGCCGCGCUUUCAUUUUGUUUGCCUGCCCUUUCAUAGAGGGUUUCUU .((.((..((((..((((.((((..(((((((((((((((...........)))))))..)))))))).)))).)))).))))((((.....)))))).)).. ( -25.20) >consensus UGAUGCUCGCAGCUCAGAGAGAAAAGAAAGUGUCAAAUUAAAUUUAAAAAGUGAUUUGCCGCGCUUUCAUUUUGUUUGCC_GCCCUUUCAUAGAGGGUCCUU_ (((.((.....))))).(((.(((((((((((((((((((...........)))))))..)))))))).)))).)))....((((((.....))))))..... (-23.50 = -23.70 + 0.20)


| Location | 13,598,306 – 13,598,414 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.35 |
| Mean single sequence MFE | -25.92 |
| Consensus MFE | -15.72 |
| Energy contribution | -16.50 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.756000 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13598306 108 + 23771897 AAGAAAGUGUCAAAUUAAAUUUAAAAAGUGAUUUGCCGCGCUUUCAUUUUGUUUGCC-GCCCUUUCAUAGAGGGUCCUU--UUUAUUUUGCCGGCUUUAAGA---------GCUUGCAAA ..(((((((((((((((...........)))))))..))))))))............-((((((.....))))))....--.....(((((.(((((...))---------))).))))) ( -28.30) >DroSec_CAF1 5162 109 + 1 AAGAAAGUGUCAAAUUAAAUUUAAAAAGUGAUUUGCCGCGCUUUCAUUUUGUUUGCC-GCCCUUUCAUAGAGGGUCCUU-UUUUAUUUUGCCGGCUUUAAAA---------GCUGGCCAA ..(((((((((((((((...........)))))))..))))))))............-((((((.....))))))....-.........(((((((.....)---------))))))... ( -30.90) >DroSim_CAF1 14540 107 + 1 AAGAAAGUGUCAAAUUAAAUUUAAAAAGUGAUUUGCCGCGCUUUCAUUUUGU-UGCC-GCCCUUUCAUAGAGGGUCCCA--UUUAUUUUGCCGGCUUUAAAA---------GCUUGCCAA ..(((((((((((((((...........)))))))..)))))))).(((((.-.(((-((((((.....))))))..((--.......))..)))..)))))---------......... ( -26.00) >DroEre_CAF1 6363 111 + 1 AAGAAAGUGUCAAAUUAAAUUUAAAAAGUGAUUUGGCGCGCUUUCAUUUUGUUUGCCUGCCCUUUCAUAGAGGGUUCUUUUUUUUUUGUGCCGGCUCUAAAA---------GCUUGCAAA ..(((((((((((((((...........)))))))))..)))))).............((((((.....)))))).............(((.((((.....)---------))).))).. ( -26.80) >DroYak_CAF1 11330 98 + 1 AAGAAAGUGUCAAAUUAAAUUUAAAAAGUGAUUUGCCGCGCUUUCAUUUUGUUUGCCUGCCCUUUCAUAGAGGGUUUCUUUUUUAUAUU----------------------GCUUGCAAA ..(((((((((((((((...........)))))))..))))))))..(((((..((..((((((.....))))))..............----------------------))..))))) ( -22.99) >DroAna_CAF1 40944 104 + 1 GGGGAAGUGUCAAAUUAAAUUUAAAAAGUGAUUUGCCGAGCUUUCAUUU-AUUUGCU------UU-------GGUUUCU--CUUGUGUCGCCGCCAUUGUCACUCCGCGGUGCUUAUUGC (.(((.(((.(((............(((.((...(((((((........-....)).------))-------))).)).--)))(((....)))..))).)))))).)............ ( -20.50) >consensus AAGAAAGUGUCAAAUUAAAUUUAAAAAGUGAUUUGCCGCGCUUUCAUUUUGUUUGCC_GCCCUUUCAUAGAGGGUCCCU__UUUAUUUUGCCGGCUUUAAAA_________GCUUGCAAA ..(((((((((((((((...........)))))))..)))))))).............((((((.....))))))............................................. (-15.72 = -16.50 + 0.78)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:35:24 2006