
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 1,365,902 – 1,365,996 |
| Length | 94 |
| Max. P | 0.783265 |

| Location | 1,365,902 – 1,365,996 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 78.54 |
| Mean single sequence MFE | -15.87 |
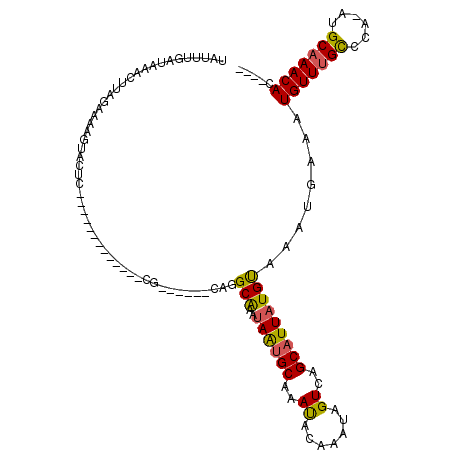
| Consensus MFE | -11.31 |
| Energy contribution | -10.87 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.783265 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
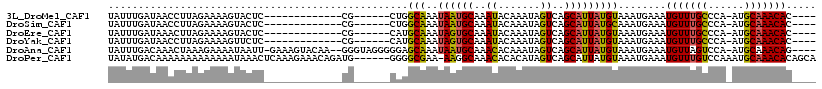
>3L_DroMel_CAF1 1365902 94 + 23771897 UAUUUGAUAACCUUAGAAAAGUACUC-------------CG------CUGGCAAAUAAUGCAAAUACAAAUAGUCAGCAUUAUGUAAAUGAAAUGUUUGCCCA-AUGCAAACAC---- ((((((((((.(((....))).....-------------.(------(((((....................)))))).)))).))))))...(((((((...-..))))))).---- ( -15.45) >DroSim_CAF1 18923 94 + 1 UAUUUGAUAACCUUAGAAAAGUACUC-------------CG------CUGGCAAAUAAUGCAAAUACAAAUAGUCAGCAUUAUGCAAAUGAAAUGUUUGCCCA-AUGCAAACAC---- ((((((((((.(((....))).....-------------.(------(((((....................)))))).)))).))))))...(((((((...-..))))))).---- ( -17.65) >DroEre_CAF1 22286 94 + 1 UAUUUGAUAAACUUAGAAAAGUACUC-------------CG------CAUGCAAAUAGUGCAAAUACAAAUAGUCAGCAUUAUGUAAAUGAAAUGUUUGCCCA-AUGCAAACAC---- ((((((....((((....))))....-------------.(------(((.......)))).....))))))((..(((((..((((((.....))))))..)-))))..))..---- ( -17.70) >DroYak_CAF1 22962 94 + 1 UAUUUGAUAACCUUAGAAAAGUUCUC-------------CG------CAUGCAAAUAGUGCAAAUACAAAUAGUCAGCAUUAUGUAAAUGAAAUGUUUGCCCA-AUGCAAACAC---- ((((((........(((.....))).-------------.(------(((.......)))).....))))))((..(((((..((((((.....))))))..)-))))..))..---- ( -16.10) >DroAna_CAF1 22417 110 + 1 UAUUUGACAAACUAAAGAAAAUAAUU-GAAAGUACAA--GGGUAGGGGGAGCAAAUAAUGCAAACACAAAUAGUCAGCAUUAUGUAAAUGAAAUGUUAGUCCA-AUGCAAACAG---- ..((((.((...........((..((-(......)))--..)).(((..((((.(((((((..((.......))..)))))))..........))))..))).-.))))))...---- ( -13.10) >DroPer_CAF1 42676 111 + 1 UAUAUGACAAAAAAAAAAAAAUAAACUCAAAGAAACAGAUG------GGGGCGAA-AAGGCAAACACACAUAGUCAGCAUUAUGUAAAUGAAAUGUUUGUCCAAAUGCAAACACAGCA ....((((.................((((..........))------)).((...-...))...........))))((.((((....))))..(((((((......)))))))..)). ( -15.20) >consensus UAUUUGAUAAACUUAGAAAAGUACUC_____________CG______CAGGCAAAUAAUGCAAAUACAAAUAGUCAGCAUUAUGUAAAUGAAAUGUUUGCCCA_AUGCAAACAC____ ..................................................(((..((((((..((.......))..)))))))))........(((((((......)))))))..... (-11.31 = -10.87 + -0.44)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:43:11 2006