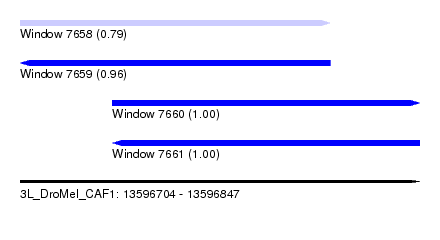
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,596,704 – 13,596,847 |
| Length | 143 |
| Max. P | 0.999399 |

| Location | 13,596,704 – 13,596,815 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.36 |
| Mean single sequence MFE | -32.81 |
| Consensus MFE | -24.64 |
| Energy contribution | -24.69 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.53 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.788899 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
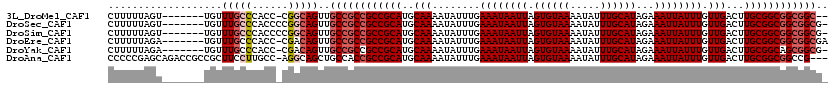
>3L_DroMel_CAF1 13596704 111 + 23771897 AAUUAAGGCGGCCAAGAAUUUUUGGGCAUCCCC---------GCCGCCGCCGCAAGUCAACAAAUAAUUUCUAUGCAAAUAUUUUACACUAAUUAUUUCAAAUAUUUUGCAUGCGGCGGC ......((((((((((....)))))......))---------)))((((((((..(....)...........((((((((((((...............))))).))))))))))))))) ( -34.26) >DroSec_CAF1 3526 114 + 1 AAUUAGGGCGGCCAAGAAUUUUUGGGCAUCCCCAG------CGCCGCCGCCGCAAGUCAACAAAUAAUUUCUAUGCAAAUAUUUUACACUAAUUAUUUCAAAUAUUUUGCAUGCGGCGGC .....(((.(.(((((....))))).)...)))..------.(((((((((....)................((((((((((((...............))))).))))))))))))))) ( -33.66) >DroSim_CAF1 12899 114 + 1 AAUUAAAGCGGCCAAGAAUUUUUGGGCAUCCCCAG------CGCCGCCGCCGCAAGUCAACAAAUAAUUUCUAUGCAAAUAUUUUACACUAAUUAUUUCAAAUAUUUUGCAUGCGGCGGC .........(.(((((....))))).)........------.(((((((((....)................((((((((((((...............))))).))))))))))))))) ( -29.86) >DroEre_CAF1 4714 120 + 1 AAUUAAGGCGGCCAAGAAUUUUUCGGCAUCCCCGCUGCCGUCGCCGCCGCCGCAAGUCAACAAAUAAUUUCUAUGCAAAUAUUUUACACUAAUUAUUUCAAAUAUUUUGCAUGCGGCGGC ......((((((...((.....))((((.......))))))))))((((((((..(....)...........((((((((((((...............))))).))))))))))))))) ( -38.46) >DroYak_CAF1 9657 114 + 1 AAUUAAGGCGGCCAAGAAUUUUUCGGCUUCCCCGC------CGCCGCUGCCGCAAGUCAACAAAUAAUUUCUAUGCAAAUAUUUUACACUAAUUAUUUCAAAUAUUUUGCAUGCGGCGGC ......((((((...((.....))((.....))))------))))((((((((..(....)...........((((((((((((...............))))).))))))))))))))) ( -34.16) >DroAna_CAF1 39246 107 + 1 AAUUAAGGCUGCACAUAUUUUUUCGGCC--CC-----------CGGCCGCCGCAAGUCAACAAAUAAUUUCUAUGCAAAUAUUUUACACUAAUUAUUUCAAAUAUUUUGCAUGCGGCGGU ......(((((............)))))--..-----------..((((((((..(....)...........((((((((((((...............))))).))))))))))))))) ( -26.46) >consensus AAUUAAGGCGGCCAAGAAUUUUUCGGCAUCCCC________CGCCGCCGCCGCAAGUCAACAAAUAAUUUCUAUGCAAAUAUUUUACACUAAUUAUUUCAAAUAUUUUGCAUGCGGCGGC ......((..(((...........)))...)).............((((((((..(....)...........((((((((((((...............))))).))))))))))))))) (-24.64 = -24.69 + 0.06)



| Location | 13,596,704 – 13,596,815 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.36 |
| Mean single sequence MFE | -38.17 |
| Consensus MFE | -30.38 |
| Energy contribution | -31.11 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.57 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.53 |
| SVM RNA-class probability | 0.961754 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13596704 111 - 23771897 GCCGCCGCAUGCAAAAUAUUUGAAAUAAUUAGUGUAAAAUAUUUGCAUAGAAAUUAUUUGUUGACUUGCGGCGGCGGC---------GGGGAUGCCCAAAAAUUCUUGGCCGCCUUAAUU (((((((((..(((........((((((((.((((((.....))))))...)))))))).)))...)))))))))(((---------((......((((......)))))))))...... ( -38.00) >DroSec_CAF1 3526 114 - 1 GCCGCCGCAUGCAAAAUAUUUGAAAUAAUUAGUGUAAAAUAUUUGCAUAGAAAUUAUUUGUUGACUUGCGGCGGCGGCG------CUGGGGAUGCCCAAAAAUUCUUGGCCGCCCUAAUU (((((((((..(((........((((((((.((((((.....))))))...)))))))).)))...)))))))))((((------(..(((((........)))))..).))))...... ( -42.20) >DroSim_CAF1 12899 114 - 1 GCCGCCGCAUGCAAAAUAUUUGAAAUAAUUAGUGUAAAAUAUUUGCAUAGAAAUUAUUUGUUGACUUGCGGCGGCGGCG------CUGGGGAUGCCCAAAAAUUCUUGGCCGCUUUAAUU (((((((((..(((........((((((((.((((((.....))))))...)))))))).)))...)))))))))((((------(..(((((........)))))..).))))...... ( -39.80) >DroEre_CAF1 4714 120 - 1 GCCGCCGCAUGCAAAAUAUUUGAAAUAAUUAGUGUAAAAUAUUUGCAUAGAAAUUAUUUGUUGACUUGCGGCGGCGGCGACGGCAGCGGGGAUGCCGAAAAAUUCUUGGCCGCCUUAAUU ((((((((.(((((........((((((((.((((((.....))))))...))))))))......)))))))))))))...(((.((.((((...........)))).)).)))...... ( -41.44) >DroYak_CAF1 9657 114 - 1 GCCGCCGCAUGCAAAAUAUUUGAAAUAAUUAGUGUAAAAUAUUUGCAUAGAAAUUAUUUGUUGACUUGCGGCAGCGGCG------GCGGGGAAGCCGAAAAAUUCUUGGCCGCCUUAAUU (((((((((((((((.(((((...((.....))...))))))))))))..........(((((.....)))))))))))------))((((..(((((.......)))))..)))).... ( -40.60) >DroAna_CAF1 39246 107 - 1 ACCGCCGCAUGCAAAAUAUUUGAAAUAAUUAGUGUAAAAUAUUUGCAUAGAAAUUAUUUGUUGACUUGCGGCGGCCG-----------GG--GGCCGAAAAAAUAUGUGCAGCCUUAAUU .((((((((..(((........((((((((.((((((.....))))))...)))))))).)))...))))))))...-----------((--(((.(............).))))).... ( -27.00) >consensus GCCGCCGCAUGCAAAAUAUUUGAAAUAAUUAGUGUAAAAUAUUUGCAUAGAAAUUAUUUGUUGACUUGCGGCGGCGGCG________GGGGAUGCCCAAAAAUUCUUGGCCGCCUUAAUU ((((((((.(((((........((((((((.((((((.....))))))...))))))))......))))))))))))).........((((..(((...........)))..)))).... (-30.38 = -31.11 + 0.72)


| Location | 13,596,737 – 13,596,847 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.66 |
| Mean single sequence MFE | -38.19 |
| Consensus MFE | -33.66 |
| Energy contribution | -33.83 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.11 |
| Mean z-score | -4.15 |
| Structure conservation index | 0.88 |
| SVM decision value | 3.57 |
| SVM RNA-class probability | 0.999399 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13596737 110 + 23771897 --GCCGCCGCCGCAAGUCAACAAAUAAUUUCUAUGCAAAUAUUUUACACUAAUUAUUUCAAAUAUUUUGCAUGCGGCGGCGGCAACUGCCG-GGUGGGCAAACA-------ACUAAAAAG --(((((((((((..(....)...........((((((((((((...............))))).))))))))))))))))))...((((.-....))))....-------......... ( -38.46) >DroSec_CAF1 3561 112 + 1 -CGCCGCCGCCGCAAGUCAACAAAUAAUUUCUAUGCAAAUAUUUUACACUAAUUAUUUCAAAUAUUUUGCAUGCGGCGGCGGCAACUGCCGGGGUGGGCAAACA-------ACUAAAAAG -.(((((((((((..(....)...........((((((((((((...............))))).))))))))))))))))))...((((......))))....-------......... ( -38.96) >DroSim_CAF1 12934 112 + 1 -CGCCGCCGCCGCAAGUCAACAAAUAAUUUCUAUGCAAAUAUUUUACACUAAUUAUUUCAAAUAUUUUGCAUGCGGCGGCGGCAACUGCCGGGGUGGGCAAACA-------ACUAAAAAG -.(((((((((((..(....)...........((((((((((((...............))))).))))))))))))))))))...((((......))))....-------......... ( -38.96) >DroEre_CAF1 4754 112 + 1 UCGCCGCCGCCGCAAGUCAACAAAUAAUUUCUAUGCAAAUAUUUUACACUAAUUAUUUCAAAUAUUUUGCAUGCGGCGGCGGCAACUGUCG-GGUGGGCAAACA-------UCUAAAAAG ..(((((((((((..(....)...........((((((((((((...............))))).)))))))))))))))))).......(-((((......))-------)))...... ( -38.16) >DroYak_CAF1 9692 111 + 1 -CGCCGCUGCCGCAAGUCAACAAAUAAUUUCUAUGCAAAUAUUUUACACUAAUUAUUUCAAAUAUUUUGCAUGCGGCGGCGGCAACUGUCG-GGUGGGCAAACA-------UCUAAAAAG -.(((((((((((..(....)...........((((((((((((...............))))).)))))))))))))))))).......(-((((......))-------)))...... ( -35.96) >DroAna_CAF1 39276 116 + 1 ---CGGCCGCCGCAAGUCAACAAAUAAUUUCUAUGCAAAUAUUUUACACUAAUUAUUUCAAAUAUUUUGCAUGCGGCGGUGGCAGCUGCCU-GGCAAGGAAGCGGCGGUCUGCUCGGGGG ---((((((((((..(....)...........((((((((((((...............))))).)))))))))))))))((((((((((.-.((......))))))).))))))).... ( -38.66) >consensus _CGCCGCCGCCGCAAGUCAACAAAUAAUUUCUAUGCAAAUAUUUUACACUAAUUAUUUCAAAUAUUUUGCAUGCGGCGGCGGCAACUGCCG_GGUGGGCAAACA_______ACUAAAAAG ..(((((((((((..(....)...........((((((((((((...............))))).))))))))))))))))))...((((......)))).................... (-33.66 = -33.83 + 0.17)

| Location | 13,596,737 – 13,596,847 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.66 |
| Mean single sequence MFE | -37.47 |
| Consensus MFE | -33.26 |
| Energy contribution | -34.28 |
| Covariance contribution | 1.03 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.27 |
| Structure conservation index | 0.89 |
| SVM decision value | 3.13 |
| SVM RNA-class probability | 0.998540 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13596737 110 - 23771897 CUUUUUAGU-------UGUUUGCCCACC-CGGCAGUUGCCGCCGCCGCAUGCAAAAUAUUUGAAAUAAUUAGUGUAAAAUAUUUGCAUAGAAAUUAUUUGUUGACUUGCGGCGGCGGC-- .........-------...(((((....-.)))))..((((((((((((..(((........((((((((.((((((.....))))))...)))))))).)))...))))))))))))-- ( -39.50) >DroSec_CAF1 3561 112 - 1 CUUUUUAGU-------UGUUUGCCCACCCCGGCAGUUGCCGCCGCCGCAUGCAAAAUAUUUGAAAUAAUUAGUGUAAAAUAUUUGCAUAGAAAUUAUUUGUUGACUUGCGGCGGCGGCG- .........-------...(((((......)))))..((((((((((((..(((........((((((((.((((((.....))))))...)))))))).)))...)))))))))))).- ( -41.40) >DroSim_CAF1 12934 112 - 1 CUUUUUAGU-------UGUUUGCCCACCCCGGCAGUUGCCGCCGCCGCAUGCAAAAUAUUUGAAAUAAUUAGUGUAAAAUAUUUGCAUAGAAAUUAUUUGUUGACUUGCGGCGGCGGCG- .........-------...(((((......)))))..((((((((((((..(((........((((((((.((((((.....))))))...)))))))).)))...)))))))))))).- ( -41.40) >DroEre_CAF1 4754 112 - 1 CUUUUUAGA-------UGUUUGCCCACC-CGACAGUUGCCGCCGCCGCAUGCAAAAUAUUUGAAAUAAUUAGUGUAAAAUAUUUGCAUAGAAAUUAUUUGUUGACUUGCGGCGGCGGCGA .........-------((((.(.....)-.)))).((((((((((((((..(((........((((((((.((((((.....))))))...)))))))).)))...)))))))))))))) ( -37.10) >DroYak_CAF1 9692 111 - 1 CUUUUUAGA-------UGUUUGCCCACC-CGACAGUUGCCGCCGCCGCAUGCAAAAUAUUUGAAAUAAUUAGUGUAAAAUAUUUGCAUAGAAAUUAUUUGUUGACUUGCGGCAGCGGCG- .........-------((((.(.....)-.))))...(((((.((((((..(((........((((((((.((((((.....))))))...)))))))).)))...)))))).))))).- ( -30.30) >DroAna_CAF1 39276 116 - 1 CCCCCGAGCAGACCGCCGCUUCCUUGCC-AGGCAGCUGCCACCGCCGCAUGCAAAAUAUUUGAAAUAAUUAGUGUAAAAUAUUUGCAUAGAAAUUAUUUGUUGACUUGCGGCGGCCG--- .....(.((((.(.(((((......)).-.))).)))))).((((((((..(((........((((((((.((((((.....))))))...)))))))).)))...))))))))...--- ( -35.10) >consensus CUUUUUAGU_______UGUUUGCCCACC_CGGCAGUUGCCGCCGCCGCAUGCAAAAUAUUUGAAAUAAUUAGUGUAAAAUAUUUGCAUAGAAAUUAUUUGUUGACUUGCGGCGGCGGCG_ ...................(((((......)))))..((((((((((((..(((........((((((((.((((((.....))))))...)))))))).)))...)))))))))))).. (-33.26 = -34.28 + 1.03)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:35:17 2006