
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
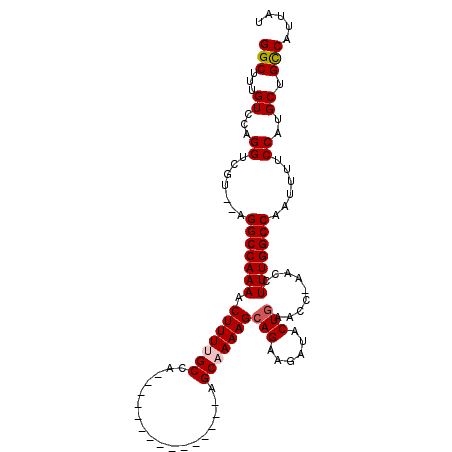
| Location | 13,595,185 – 13,595,317 |
| Length | 132 |
| Max. P | 0.826537 |

| Location | 13,595,185 – 13,595,277 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 87.00 |
| Mean single sequence MFE | -24.78 |
| Consensus MFE | -20.13 |
| Energy contribution | -20.57 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.826537 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13595185 92 - 23771897 GGCUUUGUCCAGGUCGU--AGGCCAAAACUUUUGCCA---------------AGCAAAAGCAGAAGAUACUAAACCGAACCUUUGGCCAAUUUUCCAUGCUGCCAUUAU (((...((...((..((--.(((((((.(((((((..---------------.......)))))))...(......)....))))))).))...))..)).)))..... ( -21.80) >DroSec_CAF1 2050 92 - 1 GGCUUUGUCCAGGUCGU--AGGCCAAAACUUUUGCCA---------------AGCCAAAGCAGAAGAUACUGAACCAAACCUUUGGCCAAUUUUCCAUGCUGCCAUUAU (((...((...((..((--.(((((((.(((((((..---------------.......))))))).....(........)))))))).))...))..)).)))..... ( -21.30) >DroSim_CAF1 11408 92 - 1 GGCUUUGUCCAGGUCGU--AGGCCAAAACUUUUGCCA---------------AGCAAAAGCAGAAGAUACUGAACCAAACCUUUGGCCAAUUUUCCAUGCUGCCAUUAU (((...((...((..((--.(((((((.(((((((..---------------.)))))))(((......))).........))))))).))...))..)).)))..... ( -24.30) >DroEre_CAF1 3263 89 - 1 GGCUUUGUCCAGGUCGU--AGGCCAAAACUUUUGC-AAGC------------AGCAAAAGCAGAAGAUACUGA-----ACCUUUGGCCAAUUUUCCAUGCUGCCAUUAU (((...((...((..((--.(((((((.(((((((-....------------.)))))))(((......))).-----...))))))).))...))..)).)))..... ( -25.90) >DroYak_CAF1 8070 104 - 1 GGCUUUGUCCAGGUCGUGUUGGCCAAAACUUUUGCCAAGCAUGCCAGGCAAAAGCUAAAGCAGAAGAUACUGA-----ACCUUUGGCCAAUUUUCCAUGCUGUCAUUAU (((...((...((....((((((((((.((((((((..(....)..))))))))......(((......))).-----...))))))))))...))..)).)))..... ( -30.60) >consensus GGCUUUGUCCAGGUCGU__AGGCCAAAACUUUUGCCA_______________AGCAAAAGCAGAAGAUACUGAACC_AACCUUUGGCCAAUUUUCCAUGCUGCCAUUAU (((...((...((.......(((((((.(((((((..................)))))))(((......))).........)))))))......))..)).)))..... (-20.13 = -20.57 + 0.44)

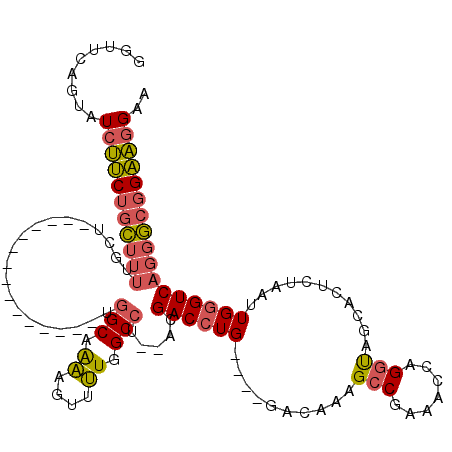
| Location | 13,595,218 – 13,595,317 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.92 |
| Mean single sequence MFE | -34.16 |
| Consensus MFE | -21.07 |
| Energy contribution | -21.18 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.543897 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13595218 99 + 23771897 GGUUUAGUAUCUUCUGCUUUUGCU---------------UGGCAAAAGUUUUGGCCU--ACGACCUG----GACAAAGCCGAAACCAGGUAGCACUCUAAUUGGGUCAGGGCGGAAGGAA .........(((((((((((((((---------------(((.....((((((.((.--.......)----).)))))).....)))))))(.((((.....)))))))))))))))).. ( -31.50) >DroSec_CAF1 2083 99 + 1 GGUUCAGUAUCUUCUGCUUUGGCU---------------UGGCAAAAGUUUUGGCCU--ACGACCUG----GACAAAGCCGAAAUCAGGUAGCACUCUAAUUGGGUCAGGGCGGAAGGAA .........(((((((((((((((---------------(((.....((((((.((.--.......)----).)))))).....))))))...((((.....)))))))))))))))).. ( -31.10) >DroSim_CAF1 11441 99 + 1 GGUUCAGUAUCUUCUGCUUUUGCU---------------UGGCAAAAGUUUUGGCCU--ACGACCUG----GACAAAGCCGAAACCAGGUAGCACUCUAAUUGGGUCAGGGCGGAAGGAA .........(((((((((((((((---------------(((.....((((((.((.--.......)----).)))))).....)))))))(.((((.....)))))))))))))))).. ( -31.50) >DroEre_CAF1 3294 98 + 1 ---UCAGUAUCUUCUGCUUUUGCU------------GCUU-GCAAAAGUUUUGGCCU--ACGACCUG----GACAAAGCCGAAACCAGGUAGCACUCUAAUUGGGUCAGGGCGGAAGGAA ---......(((((((((((.(((------------((((-(.....((((((((..--........----......))))))))))))))))((((.....)))).))))))))))).. ( -33.89) >DroYak_CAF1 8101 113 + 1 ---UCAGUAUCUUCUGCUUUAGCUUUUGCCUGGCAUGCUUGGCAAAAGUUUUGGCCAACACGACCUG----GACAAAGCCGAAACCAGGUAGCACUCUAAUUGGGUCAGGGCGGAAGGAA ---......(((((((((((((((((((((.((....)).))))))))))..((((......(((((----(............))))))....(.......)))))))))))))))).. ( -39.80) >DroAna_CAF1 38005 103 + 1 GGCGCAGUCUAACCACG--------UUUCCCGGC------GGCGGCGG--GUGGCCGACAAGACAUGUGUGUCCAUGGGCAGCACCUGGCAGCG-UCUAAUUGGGUCAGGCCGGGUGGAG .(((((((((.....((--------(......))------).((((..--...))))...)))).)))))(((....)))..((((((((...(-.((.....)).)..))))))))... ( -37.20) >consensus GGUUCAGUAUCUUCUGCUUUUGCU_______________UGGCAAAAGUUUUGGCCU__ACGACCUG____GACAAAGCCGAAACCAGGUAGCACUCUAAUUGGGUCAGGGCGGAAGGAA .........(((((((((((....................(((.((....)).))).....((((((..........(((.......)))...........))))))))))))))))).. (-21.07 = -21.18 + 0.12)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:35:13 2006