
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,587,983 – 13,588,128 |
| Length | 145 |
| Max. P | 0.997866 |

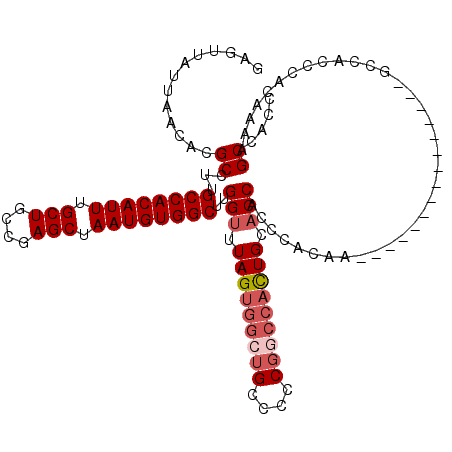
| Location | 13,587,983 – 13,588,088 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.31 |
| Mean single sequence MFE | -34.50 |
| Consensus MFE | -27.64 |
| Energy contribution | -29.20 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.95 |
| Structure conservation index | 0.80 |
| SVM decision value | 2.95 |
| SVM RNA-class probability | 0.997866 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13587983 105 - 23771897 GAGUUAUUAACACGCCUUGCCACAUUUGCUGCCGAGCUAAUGUGGCUUGGUUUAGUGGCUGCCCCCGGCCACUGCACCACCCACAA---------------GCCACCCACAAAAGCCACC .............((...((((((((.(((....))).)))))))).((((.(((((((((....))))))))).)))).......---------------))................. ( -35.50) >DroSim_CAF1 3778 105 - 1 GAGUUAUUAACACGCCUUGCCACAUUUGCUGCCGAGCUAAUGUGGCUUGGUUUAGUGGCUGCCCCCGGCCACUGCACCACCCACAA---------------GCCACCCACAAAAGCCACC .............((...((((((((.(((....))).)))))))).((((.(((((((((....))))))))).)))).......---------------))................. ( -35.50) >DroYak_CAF1 738 120 - 1 GAGUUAUUAACACGCCUUGCCACAUUUGCUGCCGAGCUAAUGUGGCUUGGUUUAGUGGCUGCCCCCGACCACUGCACCACCCACAAAAGCCACCCACAAAAACCACCCACAAGAGCCACC .............(((((((((((((.(((....))).)))))))).((((.((((((.((....)).)))))).))))...............................))).)).... ( -30.90) >DroAna_CAF1 31008 108 - 1 GAGUUAUUAACACGCUUUGCCACAUUUGCUGCCAAGCUAAUGUGGCCAGGUUUAGUGGCUGCCUCC-----UUGCCCCAGCCACCC-AGCCACCCAGCU-UGCCACC-----GAGCCACC ..................((((((((.(((....))).))))))))..((((..(((((((.....-----......)))))))..-)))).....(((-((....)-----)))).... ( -36.10) >consensus GAGUUAUUAACACGCCUUGCCACAUUUGCUGCCGAGCUAAUGUGGCUUGGUUUAGUGGCUGCCCCCGGCCACUGCACCACCCACAA_______________GCCACCCACAAAAGCCACC .............((...((((((((.(((....))).))))))))..(((.(((((((((....))))))))).)))....................................)).... (-27.64 = -29.20 + 1.56)


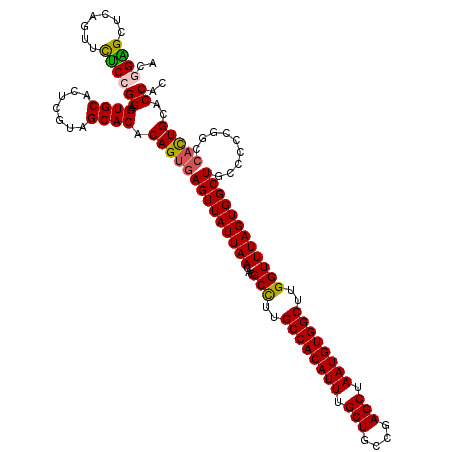
| Location | 13,588,008 – 13,588,128 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.61 |
| Mean single sequence MFE | -43.20 |
| Consensus MFE | -34.35 |
| Energy contribution | -34.47 |
| Covariance contribution | 0.13 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.60 |
| SVM RNA-class probability | 0.966898 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13588008 120 - 23771897 GCUGAGCUCAGUUCUCCGGAGUGCACUCGUAGCACACAGUGAGUUAUUAACACGCCUUGCCACAUUUGCUGCCGAGCUAAUGUGGCUUGGUUUAGUGGCUGCCCCCGGCCACUGCACCAC ((((....)))).....((.((((.......)))).((((((((((((((...(((..((((((((.(((....))).))))))))..))))))))))))(((...))))))))..)).. ( -45.80) >DroSim_CAF1 3803 120 - 1 ACGGGGCUCAGUUCUCCGGAGUGCACUCGUAGCACACAGUGAGUUAUUAACACGCCUUGCCACAUUUGCUGCCGAGCUAAUGUGGCUUGGUUUAGUGGCUGCCCCCGGCCACUGCACCAC ..(((((.............((((.......))))...(((.........))))))))((((((((.(((....))).)))))))).((((.(((((((((....))))))))).)))). ( -47.40) >DroYak_CAF1 778 120 - 1 ACAGAGCUCAGCUCUCCGGAGUGCACUCGUAGCACACAGUGAGUUAUUAACACGCCUUGCCACAUUUGCUGCCGAGCUAAUGUGGCUUGGUUUAGUGGCUGCCCCCGACCACUGCACCAC ..(((((...)))))..((.((((.......)))).((((((((((((((...(((..((((((((.(((....))).))))))))..)))))))))))).........)))))..)).. ( -42.70) >DroAna_CAF1 31041 97 - 1 CCGGUGC-----UUGCCGGAG-------------CACAGUGAGUUAUUAACACGCUUUGCCACAUUUGCUGCCAAGCUAAUGUGGCCAGGUUUAGUGGCUGCCUCC-----UUGCCCCAG (((((..-----..))))).(-------------((.((..(((((((((...((((.((((((((.(((....))).)))))))).)))))))))))))..))..-----.)))..... ( -36.90) >consensus ACGGAGCUCAGUUCUCCGGAGUGCACUCGUAGCACACAGUGAGUUAUUAACACGCCUUGCCACAUUUGCUGCCGAGCUAAUGUGGCUUGGUUUAGUGGCUGCCCCCGGCCACUGCACCAC ..((((.......))))((.((((.......)))).((((((((((((((...(((..((((((((.(((....))).))))))))..)))))))))))).........)))))..)).. (-34.35 = -34.47 + 0.13)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:35:05 2006