
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,585,010 – 13,585,207 |
| Length | 197 |
| Max. P | 0.998486 |

| Location | 13,585,010 – 13,585,129 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 73.26 |
| Mean single sequence MFE | -38.37 |
| Consensus MFE | -22.08 |
| Energy contribution | -25.87 |
| Covariance contribution | 3.79 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.58 |
| SVM decision value | 1.95 |
| SVM RNA-class probability | 0.983580 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13585010 119 + 23771897 AGCAGCAAACUUGUCUAAGCUUUCGACCAAGACAGAUAGAUAGGGUUGCGAAGA-GCCUUGGGGGCUCGGCGAAGAGCCGAGCUCUUGCAAUUACAAUUACGCAUAGACGGGGGAAAACA .(((((...((((((((..((.((......)).)).))))))))))))).....-.(((((((((((((((.....))))))))))(((............)))....)))))....... ( -41.30) >DroSim_CAF1 789 119 + 1 AGCAGCAAACUUGUCUAAGCUUUCGACCAAGACAGAUAGAUAGGGUUGCGAAGA-GCCUUGGGGGCUCGGCGAAGAGCCGAGCUCUUGCAAUUACAAUUACGCAUAGACGGGGGAAAACA .(((((...((((((((..((.((......)).)).))))))))))))).....-.(((((((((((((((.....))))))))))(((............)))....)))))....... ( -41.30) >DroAna_CAF1 27519 103 + 1 UGCAGCGAGCUUGCCUAAGCCUCCGUCCAAGACGGACAGG-------ACGAAAAGGCCUUAGGGAACGGGCGAA-------GCCUUCGCAAUUACAACUUCGUUGGGAAGGAAGGAA--- ........((((((((((((((.(((((..........))-------)))...))).))))))...)))))...-------.(((((.((((.........)))).)))))......--- ( -32.50) >consensus AGCAGCAAACUUGUCUAAGCUUUCGACCAAGACAGAUAGAUAGGGUUGCGAAGA_GCCUUGGGGGCUCGGCGAAGAGCCGAGCUCUUGCAAUUACAAUUACGCAUAGACGGGGGAAAACA .((.((((.(((((((...((.((......)).))..))))))).))))......((...(((((((((((.....)))))))))))))............))................. (-22.08 = -25.87 + 3.79)

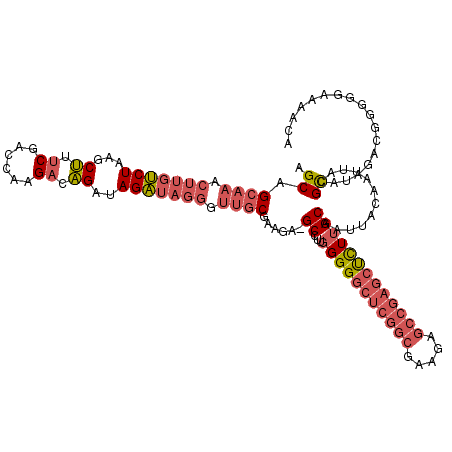
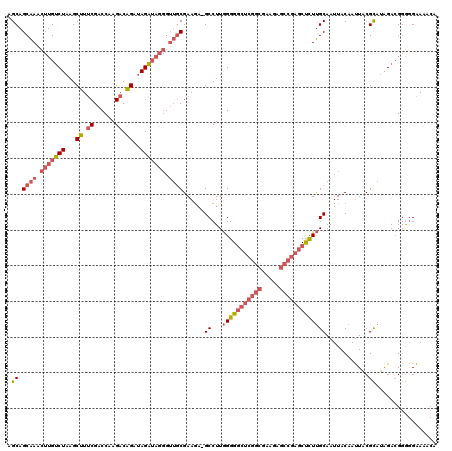
| Location | 13,585,089 – 13,585,207 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 67.32 |
| Mean single sequence MFE | -30.53 |
| Consensus MFE | -17.55 |
| Energy contribution | -16.57 |
| Covariance contribution | -0.99 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.57 |
| SVM decision value | 3.12 |
| SVM RNA-class probability | 0.998486 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13585089 118 + 23771897 AGCUCUUGCAAUUACAAUUACGCAUAGACGGGGGAAAACAAAACAGCAUUGGAUGAUGAAGAGCUUGGCCAAGAGAAACGGCCAA-CGGUAAUUGUA-AUGGCCGACAAAGUUGGCAAAA .((....)).(((((((((((((......(........).......((((....))))....))((((((.........))))))-..)))))))))-)).((((((...)))))).... ( -31.00) >DroSim_CAF1 868 118 + 1 AGCUCUUGCAAUUACAAUUACGCAUAGACGGGGGAAAACAAAACAGCAGUGGAUGAUGAAGAGCUUGGCCAAGAGAAACGGCCAA-CGGUAAUUGUA-AUGGCCGACAAAGUUGGCAAAA .((....)).(((((((((((.(((..((...(..........)....))..))).......(.((((((.........))))))-).)))))))))-)).((((((...)))))).... ( -30.30) >DroAna_CAF1 27586 95 + 1 -GCCUUCGCAAUUACAACUUCGUUGGGAAGGAAGGAA----------------------GGUGGGUGGCCA-GAC-AAGGGCCACUUGCUAAUCGUAAAUGGUCAACAAUGUAGGUGAGG -..((((((...((((.((((.....))))...((..----------------------((..(((((((.-...-...)))))))..))..))...............)))).)))))) ( -30.30) >consensus AGCUCUUGCAAUUACAAUUACGCAUAGACGGGGGAAAACAAAACAGCA_UGGAUGAUGAAGAGCUUGGCCAAGAGAAACGGCCAA_CGGUAAUUGUA_AUGGCCGACAAAGUUGGCAAAA .......((((((((..(((....)))...................................(.((((((.........)))))).).)))))))).....((((((...)))))).... (-17.55 = -16.57 + -0.99)



| Location | 13,585,089 – 13,585,207 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 67.32 |
| Mean single sequence MFE | -25.19 |
| Consensus MFE | -15.12 |
| Energy contribution | -15.13 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.60 |
| SVM decision value | 2.15 |
| SVM RNA-class probability | 0.989121 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13585089 118 - 23771897 UUUUGCCAACUUUGUCGGCCAU-UACAAUUACCG-UUGGCCGUUUCUCUUGGCCAAGCUCUUCAUCAUCCAAUGCUGUUUUGUUUUCCCCCGUCUAUGCGUAAUUGUAAUUGCAAGAGCU ....(((((......((((((.-...........-.))))))......)))))..(((((((((.((....((((.....((........)).....))))...))....)).))))))) ( -27.42) >DroSim_CAF1 868 118 - 1 UUUUGCCAACUUUGUCGGCCAU-UACAAUUACCG-UUGGCCGUUUCUCUUGGCCAAGCUCUUCAUCAUCCACUGCUGUUUUGUUUUCCCCCGUCUAUGCGUAAUUGUAAUUGCAAGAGCU .........((((....((.((-(((((((((((-(((((((.......))))))(((...............)))...................))).))))))))))).)).)))).. ( -27.16) >DroAna_CAF1 27586 95 - 1 CCUCACCUACAUUGUUGACCAUUUACGAUUAGCAAGUGGCCCUU-GUC-UGGCCACCCACC----------------------UUCCUUCCUUCCCAACGAAGUUGUAAUUGCGAAGGC- ...........(((((((.(......).)))))))((((((...-...-.))))))...((----------------------(((....((((.....))))..((....))))))).- ( -21.00) >consensus UUUUGCCAACUUUGUCGGCCAU_UACAAUUACCG_UUGGCCGUUUCUCUUGGCCAAGCUCUUCAUCAUCCA_UGCUGUUUUGUUUUCCCCCGUCUAUGCGUAAUUGUAAUUGCAAGAGCU ((((((...(......)......(((((((((...(((((((.......)))))))...................................((....)))))))))))...))))))... (-15.12 = -15.13 + 0.01)

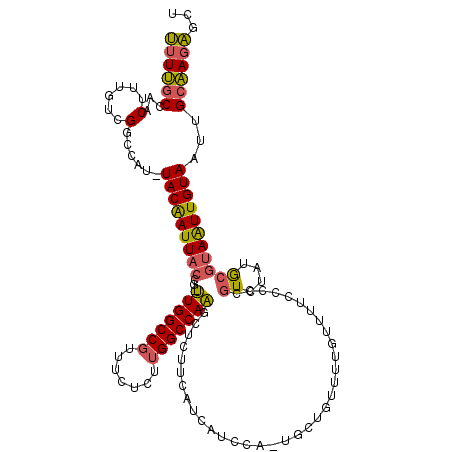
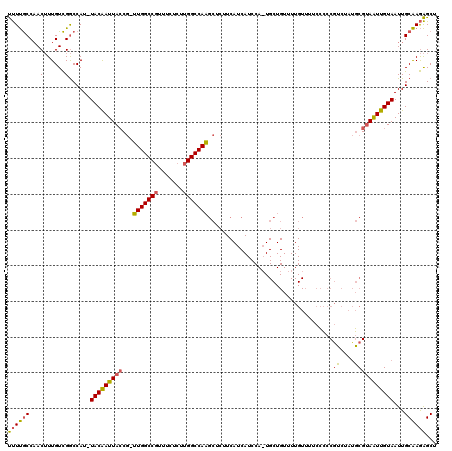
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:35:01 2006