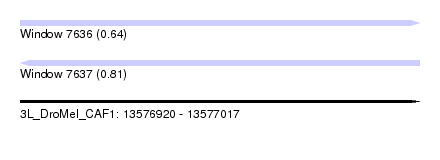
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,576,920 – 13,577,017 |
| Length | 97 |
| Max. P | 0.810290 |

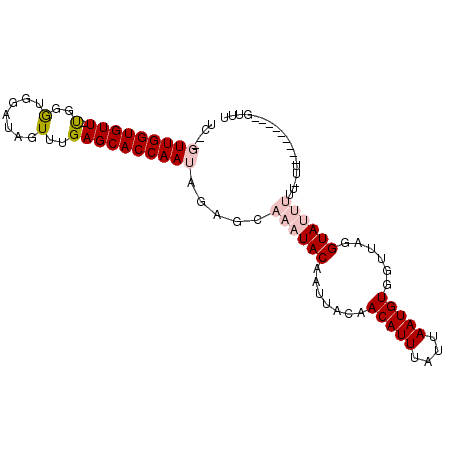
| Location | 13,576,920 – 13,577,017 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 82.54 |
| Mean single sequence MFE | -21.10 |
| Consensus MFE | -12.91 |
| Energy contribution | -13.89 |
| Covariance contribution | 0.97 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.639309 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13576920 97 + 23771897 UC--GUUGGUGUU-UGGGUGGAUAGUUUGAGCACCAAUAGAGCAAAUACAAUUACAACAUUUAUUAAUGUGGUUAGGUAUUUUUUUUUUUUUUUUGGUUU ..--(((((((((-..((.......))..)))))))))((((.((((((....((.(((((....))))).))...)))))).))))............. ( -21.50) >DroPse_CAF1 66776 80 + 1 UUUUUUUGGUGUUUCGGGUGGAUAUUUUGAGCACCAAUAGAGCAAAUACAGUUACAACAUUUAUUAAUGUGG---GGUAACGG----------------- ((((.(((((((((..(((....)))..))))))))).))))........(((((.(((((....)))))..---.)))))..----------------- ( -20.60) >DroSim_CAF1 44569 89 + 1 UC--GUUGGUGUU-UGGGUGGAUAGUUUGAGCACCAAUAGAGCAAAUACAAUUACAACAUUUAUUAAUGUGGUUAGGUAUUUUU-UUU-------UGUUU ..--(((((((((-..((.......))..)))))))))((((.((((((....((.(((((....))))).))...)))))).)-)))-------..... ( -21.50) >DroEre_CAF1 59785 88 + 1 UC--GUUGGUGUU-UGGGUGGAUAGUUUGAGCACCAAUAGAGCAAAUACAAUUACAACAUUUAUUAAUGUGGUUAGGUAUUUUUUUU---------GUUU ..--(((((((((-..((.......))..)))))))))((((.((((((....((.(((((....))))).))...)))))).))))---------.... ( -22.10) >DroYak_CAF1 61209 91 + 1 UC--GUUGGUGUUUUGGAUGGAUAGUUUGAGCACCAAUAGAGCAAAUACAAUUACAACAUUUAUUAAUGUGGUUAGGUAUUUUUUUUU-------GGUUU ..--((((((((.(..(((.....)))..)))))))))((((.((((((....((.(((((....))))).))...)))))).)))).-------..... ( -20.60) >DroPer_CAF1 66427 79 + 1 UUU-UUUGGUGUUUCGGGUGGAUAUUUUGAGCACCAAUAGAGCAAAUACAGUUACAACAUUUAUUAAUGUGG---GGUAACGG----------------- (((-.(((((((((..(((....)))..))))))))).))).........(((((.(((((....)))))..---.)))))..----------------- ( -20.30) >consensus UC__GUUGGUGUU_UGGGUGGAUAGUUUGAGCACCAAUAGAGCAAAUACAAUUACAACAUUUAUUAAUGUGGUUAGGUAUUUUU_UU_________GUUU ....(((((((((.(..(.......)..)))))))))).....((((((.......(((((....)))))......)))))).................. (-12.91 = -13.89 + 0.97)

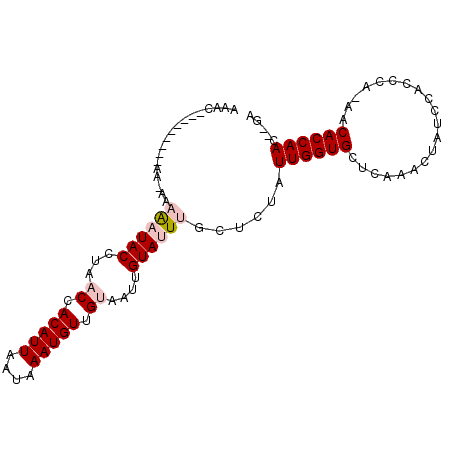
| Location | 13,576,920 – 13,577,017 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 82.54 |
| Mean single sequence MFE | -10.77 |
| Consensus MFE | -7.77 |
| Energy contribution | -8.55 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.810290 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13576920 97 - 23771897 AAACCAAAAAAAAAAAAAAAAUACCUAACCACAUUAAUAAAUGUUGUAAUUGUAUUUGCUCUAUUGGUGCUCAAACUAUCCACCCA-AACACCAAC--GA ..................((((((...((.(((((....))))).))....))))))......((((((.................-..)))))).--.. ( -10.61) >DroPse_CAF1 66776 80 - 1 -----------------CCGUUACC---CCACAUUAAUAAAUGUUGUAACUGUAUUUGCUCUAUUGGUGCUCAAAAUAUCCACCCGAAACACCAAAAAAA -----------------..(((((.---..(((((....))))).))))).............((((((.((.............))..))))))..... ( -11.12) >DroSim_CAF1 44569 89 - 1 AAACA-------AAA-AAAAAUACCUAACCACAUUAAUAAAUGUUGUAAUUGUAUUUGCUCUAUUGGUGCUCAAACUAUCCACCCA-AACACCAAC--GA .....-------...-..((((((...((.(((((....))))).))....))))))......((((((.................-..)))))).--.. ( -10.61) >DroEre_CAF1 59785 88 - 1 AAAC---------AAAAAAAAUACCUAACCACAUUAAUAAAUGUUGUAAUUGUAUUUGCUCUAUUGGUGCUCAAACUAUCCACCCA-AACACCAAC--GA ....---------.....((((((...((.(((((....))))).))....))))))......((((((.................-..)))))).--.. ( -10.61) >DroYak_CAF1 61209 91 - 1 AAACC-------AAAAAAAAAUACCUAACCACAUUAAUAAAUGUUGUAAUUGUAUUUGCUCUAUUGGUGCUCAAACUAUCCAUCCAAAACACCAAC--GA .....-------......((((((...((.(((((....))))).))....))))))......((((((....................)))))).--.. ( -10.55) >DroPer_CAF1 66427 79 - 1 -----------------CCGUUACC---CCACAUUAAUAAAUGUUGUAACUGUAUUUGCUCUAUUGGUGCUCAAAAUAUCCACCCGAAACACCAAA-AAA -----------------..(((((.---..(((((....))))).))))).............((((((.((.............))..)))))).-... ( -11.12) >consensus AAAC_________AA_AAAAAUACCUAACCACAUUAAUAAAUGUUGUAAUUGUAUUUGCUCUAUUGGUGCUCAAACUAUCCACCCA_AACACCAAC__GA ..................((((((...((.(((((....))))).))....))))))......((((((....................))))))..... ( -7.77 = -8.55 + 0.78)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:34:55 2006