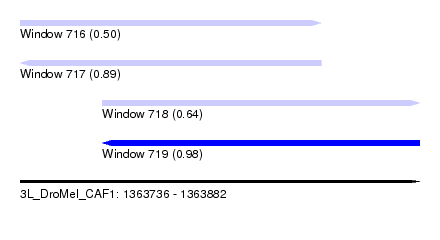
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 1,363,736 – 1,363,882 |
| Length | 146 |
| Max. P | 0.983464 |

| Location | 1,363,736 – 1,363,846 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.74 |
| Mean single sequence MFE | -31.32 |
| Consensus MFE | -27.46 |
| Energy contribution | -27.46 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.88 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1363736 110 + 23771897 -----CUC----G-UAACAAGCUUUACGAUAAGCAUACACAAGUCCCAUAUUGUCUAGUGGUUAGGAUAUCCGGCUCUCACCCGGAAGGCCCGGGUUCAAUUCCCGGUAUGGGAAAUAUG -----.((----(-(((......))))))..............(((((((.(((((........))))).((((.....((((((.....)))))).......)))))))))))...... ( -31.70) >DroSec_CAF1 19052 110 + 1 -----CUC----G-ACACAACCUUUACGAUAAGGAAACACAAGUCCCAUAUUGUCUAGUGGUUAGGAUAUCCGGCUCUCACCCGGAAGGCCCGGGUUCAAUUCCCGGUAUGGGAAAUAUG -----...----.-......((((......)))).........(((((((.(((((........))))).((((.....((((((.....)))))).......)))))))))))...... ( -30.80) >DroEre_CAF1 20080 110 + 1 -----AUC----C-UAAUAUCAUUUAUGAUAAUCAAACACAACUCCCAUAUUGUCUAGUGGUUAGGAUAUCCGGCUCUCACCCGGAAGGCCCGGGUUCAAUUCCCGGUAUGGGAAAUAUG -----...----.-...(((((....)))))............(((((((.(((((........))))).((((.....((((((.....)))))).......)))))))))))...... ( -29.60) >DroYak_CAF1 20780 110 + 1 -----CUC----G-AAAUAAUAUUUUUGAUAAACAAACACAACUCCCAUAUUGUCUAGUGGUUAGGAUAUCCGGCUCUCACCCGGAAGGCCCGGGUUCAAUUCCCGGUAUGGGAAAUAUG -----.((----(-(((......))))))..............(((((((.(((((........))))).((((.....((((((.....)))))).......)))))))))))...... ( -30.20) >DroAna_CAF1 20347 118 + 1 UUUUAAUCCUUUUGCAACACUGUUUGAGAC-GCCAUUUUCAAGUCCCAUAUUGUCUAGUGGUUAGGAUAUCCGGCUCUCACCCGGAAGGCCCGGGUUCGAUUCCCGGUAUGGGAAACCU- .............((....(((..(((((.-(((...........((((........))))...((....))))))))))..)))...))(((((.......)))))...((....)).- ( -34.30) >consensus _____CUC____G_AAACAACAUUUACGAUAAGCAAACACAAGUCCCAUAUUGUCUAGUGGUUAGGAUAUCCGGCUCUCACCCGGAAGGCCCGGGUUCAAUUCCCGGUAUGGGAAAUAUG ...........................................(((((((.(((((........))))).((((.....((((((.....)))))).......)))))))))))...... (-27.46 = -27.46 + 0.00)

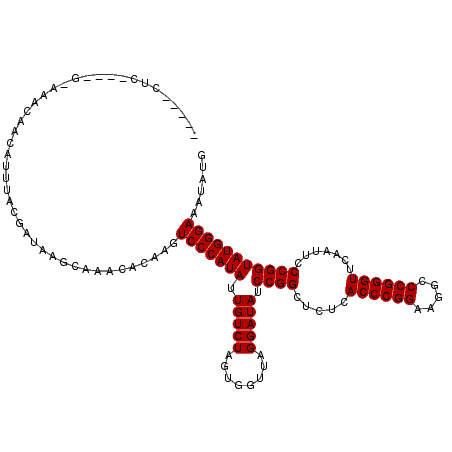
| Location | 1,363,736 – 1,363,846 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.74 |
| Mean single sequence MFE | -34.06 |
| Consensus MFE | -30.32 |
| Energy contribution | -30.60 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.887812 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1363736 110 - 23771897 CAUAUUUCCCAUACCGGGAAUUGAACCCGGGCCUUCCGGGUGAGAGCCGGAUAUCCUAACCACUAGACAAUAUGGGACUUGUGUAUGCUUAUCGUAAAGCUUGUUA-C----GAG----- ((((..(((((((((((...((..((((((.....))))))..)).)))).....(((.....)))....)))))))..))))........((((((......)))-)----)).----- ( -36.30) >DroSec_CAF1 19052 110 - 1 CAUAUUUCCCAUACCGGGAAUUGAACCCGGGCCUUCCGGGUGAGAGCCGGAUAUCCUAACCACUAGACAAUAUGGGACUUGUGUUUCCUUAUCGUAAAGGUUGUGU-C----GAG----- ((((..(((((((((((...((..((((((.....))))))..)).)))).....(((.....)))....)))))))..))))...((((......))))......-.----...----- ( -33.70) >DroEre_CAF1 20080 110 - 1 CAUAUUUCCCAUACCGGGAAUUGAACCCGGGCCUUCCGGGUGAGAGCCGGAUAUCCUAACCACUAGACAAUAUGGGAGUUGUGUUUGAUUAUCAUAAAUGAUAUUA-G----GAU----- ((((.((((((((((((...((..((((((.....))))))..)).)))).....(((.....)))....)))))))).))))((((((.((((....))))))))-)----)..----- ( -34.70) >DroYak_CAF1 20780 110 - 1 CAUAUUUCCCAUACCGGGAAUUGAACCCGGGCCUUCCGGGUGAGAGCCGGAUAUCCUAACCACUAGACAAUAUGGGAGUUGUGUUUGUUUAUCAAAAAUAUUAUUU-C----GAG----- ((((.((((((((((((...((..((((((.....))))))..)).)))).....(((.....)))....)))))))).))))((((.....))))..........-.----...----- ( -32.20) >DroAna_CAF1 20347 118 - 1 -AGGUUUCCCAUACCGGGAAUCGAACCCGGGCCUUCCGGGUGAGAGCCGGAUAUCCUAACCACUAGACAAUAUGGGACUUGAAAAUGGC-GUCUCAAACAGUGUUGCAAAAGGAUUAAAA -.((((((((.....)))))((..((((((.....))))))..)))))....(((((........((((...((((((...........-)))))).....)))).....)))))..... ( -33.42) >consensus CAUAUUUCCCAUACCGGGAAUUGAACCCGGGCCUUCCGGGUGAGAGCCGGAUAUCCUAACCACUAGACAAUAUGGGACUUGUGUUUGCUUAUCAUAAAGAUUGUUA_C____GAG_____ ((((..(((((((((((...((..((((((.....))))))..)).)))).....(((.....)))....)))))))..))))..................................... (-30.32 = -30.60 + 0.28)

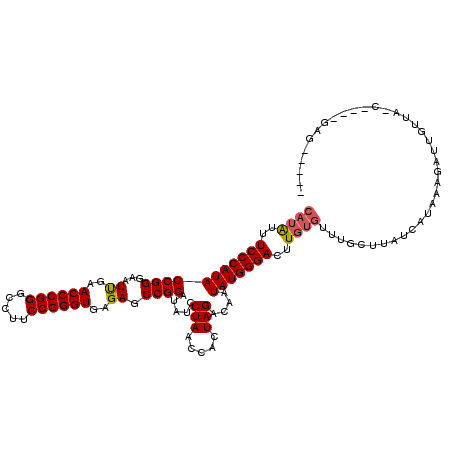
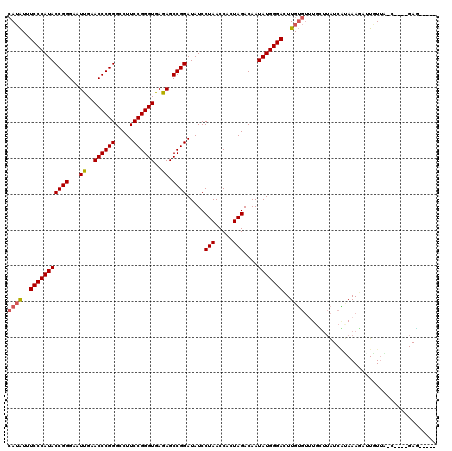
| Location | 1,363,766 – 1,363,882 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 84.21 |
| Mean single sequence MFE | -28.95 |
| Consensus MFE | -27.45 |
| Energy contribution | -27.45 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.25 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.643612 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1363766 116 + 23771897 AAGUCCCAUAUUGUCUAGUGGUUAGGAUAUCCGGCUCUCACCCGGAAGGCCCGGGUUCAAUUCCCGGUAUGGGAAAUAUGGUUUCAGAUCUUUUUUCAACA---UUAUUUUAAAAUGAA ...(((((((.(((((........))))).((((.....((((((.....)))))).......))))))))))).........................((---((.......)))).. ( -27.80) >DroEre_CAF1 20110 117 + 1 AACUCCCAUAUUGUCUAGUGGUUAGGAUAUCCGGCUCUCACCCGGAAGGCCCGGGUUCAAUUCCCGGUAUGGGAAAUAUGGAUGCUCA--UUUUUUUAUCGCGUAUAUUUUUAAAUUUA ...(((((((.(((((........))))).((((.....((((((.....)))))).......))))))))))).(((((((((....--......)))).)))))............. ( -30.80) >DroYak_CAF1 20810 116 + 1 AACUCCCAUAUUGUCUAGUGGUUAGGAUAUCCGGCUCUCACCCGGAAGGCCCGGGUUCAAUUCCCGGUAUGGGAAAUAUGAAUGCACAAUUUUUUUUAUCG---GUAUUUUAAAAUUUA ...(((((((.(((((........))))).((((.....((((((.....)))))).......))))))))))).....((((((.(.((.......)).)---))))))......... ( -28.40) >DroAna_CAF1 20386 101 + 1 AAGUCCCAUAUUGUCUAGUGGUUAGGAUAUCCGGCUCUCACCCGGAAGGCCCGGGUUCGAUUCCCGGUAUGGGAAACCU-------------UUUUUAUAA----CAUU-UUAAAUUAA ...(((((((.(((((........))))).((((.((..((((((.....))))))..))...))))))))))).....-------------.........----....-......... ( -28.80) >consensus AACUCCCAUAUUGUCUAGUGGUUAGGAUAUCCGGCUCUCACCCGGAAGGCCCGGGUUCAAUUCCCGGUAUGGGAAAUAUGGAUGCACA__UUUUUUUAUCA____UAUUUUAAAAUUAA ...(((((((.(((((........))))).((((.....((((((.....)))))).......)))))))))))............................................. (-27.45 = -27.45 + -0.00)

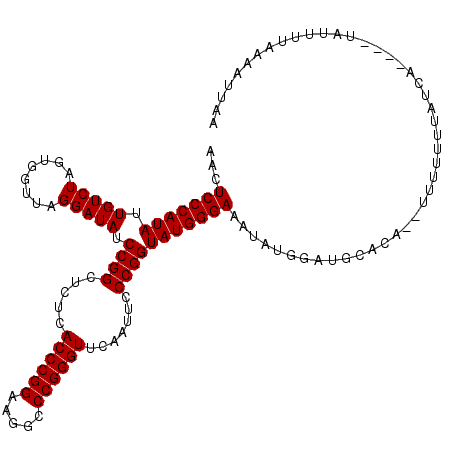
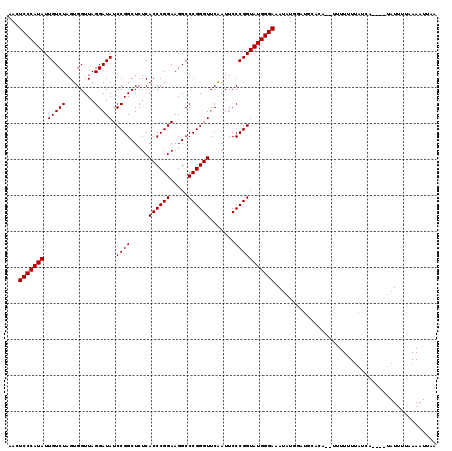
| Location | 1,363,766 – 1,363,882 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 84.21 |
| Mean single sequence MFE | -29.10 |
| Consensus MFE | -28.89 |
| Energy contribution | -28.70 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.99 |
| SVM decision value | 1.94 |
| SVM RNA-class probability | 0.983464 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1363766 116 - 23771897 UUCAUUUUAAAAUAA---UGUUGAAAAAAGAUCUGAAACCAUAUUUCCCAUACCGGGAAUUGAACCCGGGCCUUCCGGGUGAGAGCCGGAUAUCCUAACCACUAGACAAUAUGGGACUU ...............---...........................(((((((((((...((..((((((.....))))))..)).)))).....(((.....)))....)))))))... ( -28.10) >DroEre_CAF1 20110 117 - 1 UAAAUUUAAAAAUAUACGCGAUAAAAAAA--UGAGCAUCCAUAUUUCCCAUACCGGGAAUUGAACCCGGGCCUUCCGGGUGAGAGCCGGAUAUCCUAACCACUAGACAAUAUGGGAGUU .................((..........--...))........((((((((((((...((..((((((.....))))))..)).)))).....(((.....)))....)))))))).. ( -29.62) >DroYak_CAF1 20810 116 - 1 UAAAUUUUAAAAUAC---CGAUAAAAAAAAUUGUGCAUUCAUAUUUCCCAUACCGGGAAUUGAACCCGGGCCUUCCGGGUGAGAGCCGGAUAUCCUAACCACUAGACAAUAUGGGAGUU ...............---..........................((((((((((((...((..((((((.....))))))..)).)))).....(((.....)))....)))))))).. ( -28.80) >DroAna_CAF1 20386 101 - 1 UUAAUUUAA-AAUG----UUAUAAAAA-------------AGGUUUCCCAUACCGGGAAUCGAACCCGGGCCUUCCGGGUGAGAGCCGGAUAUCCUAACCACUAGACAAUAUGGGACUU .........-....----.........-------------.....(((((((((((...((..((((((.....))))))..)).)))).....(((.....)))....)))))))... ( -29.90) >consensus UAAAUUUAAAAAUA____CGAUAAAAAAA__UGUGCAUCCAUAUUUCCCAUACCGGGAAUUGAACCCGGGCCUUCCGGGUGAGAGCCGGAUAUCCUAACCACUAGACAAUAUGGGACUU .............................................(((((((((((...((..((((((.....))))))..)).)))).....(((.....)))....)))))))... (-28.89 = -28.70 + -0.19)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:43:08 2006