
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,567,549 – 13,567,672 |
| Length | 123 |
| Max. P | 0.993378 |

| Location | 13,567,549 – 13,567,652 |
|---|---|
| Length | 103 |
| Sequences | 3 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 96.09 |
| Mean single sequence MFE | -26.30 |
| Consensus MFE | -24.40 |
| Energy contribution | -24.40 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.51 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.90 |
| SVM RNA-class probability | 0.981997 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13567549 103 + 23771897 CACCAGCAUCCCCCUUUUUUCGCCCACUUCCAUGGGCCCCAGGCCAUAGGAAAUAUGGUCGAACCGAGGCCUAACUCCUUUCACCACUUUUCCCCCAAUGCCA .....................(((((......)))))....(((....(((((..((((.(((..(((......)))..)))))))..)))))......))). ( -27.90) >DroSec_CAF1 49114 101 + 1 CACCAGCAUACCCCUU-UUCCGCCCACUUCCAUGGGCCCCAGGCCAUAGGAAAUAUGGUCGAACCGAGGCCUAACUCCUUUCACCACUU-UCCCUCAAUGCCA .....((((...(((.-....(((((......)))))...))).....(((((..((((.(((..(((......)))..))))))).))-)))....)))).. ( -25.70) >DroSim_CAF1 35277 101 + 1 CACCAGCAGCCCCCUU-UUCCGCCCACUUCCAUGGGCCCCAGGCCAUAGGAAAUAUGGUCGAACCGAGGCCUAACUCCUUUCACCACUU-UCCCCCAAUGCCA ................-....(((((......)))))....(((....(((((..((((.(((..(((......)))..))))))).))-)))......))). ( -25.30) >consensus CACCAGCAUCCCCCUU_UUCCGCCCACUUCCAUGGGCCCCAGGCCAUAGGAAAUAUGGUCGAACCGAGGCCUAACUCCUUUCACCACUU_UCCCCCAAUGCCA .....................(((((......)))))....(((....(((.(..((((.(((..(((......)))..)))))))..).)))......))). (-24.40 = -24.40 + 0.00)



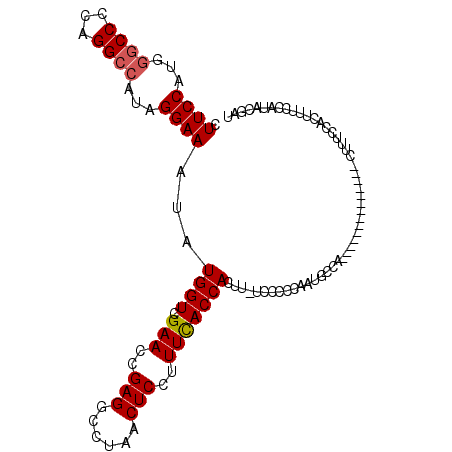
| Location | 13,567,549 – 13,567,652 |
|---|---|
| Length | 103 |
| Sequences | 3 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 96.09 |
| Mean single sequence MFE | -36.00 |
| Consensus MFE | -32.49 |
| Energy contribution | -32.27 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.880498 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13567549 103 - 23771897 UGGCAUUGGGGGAAAAGUGGUGAAAGGAGUUAGGCCUCGGUUCGACCAUAUUUCCUAUGGCCUGGGGCCCAUGGAAGUGGGCGAAAAAAGGGGGAUGCUGGUG .((((((..((((((.((((((((..(((......)))..))).))))).))))))....(((...((((((....))))))......)))..)))))).... ( -37.20) >DroSec_CAF1 49114 101 - 1 UGGCAUUGAGGGA-AAGUGGUGAAAGGAGUUAGGCCUCGGUUCGACCAUAUUUCCUAUGGCCUGGGGCCCAUGGAAGUGGGCGGAA-AAGGGGUAUGCUGGUG .(((....(((((-(.((((((((..(((......)))..))).))))).))))))(((.(((...((((((....))))))....-..))).)))))).... ( -35.30) >DroSim_CAF1 35277 101 - 1 UGGCAUUGGGGGA-AAGUGGUGAAAGGAGUUAGGCCUCGGUUCGACCAUAUUUCCUAUGGCCUGGGGCCCAUGGAAGUGGGCGGAA-AAGGGGGCUGCUGGUG ...(((..(((((-((((((((((..(((......)))..))).))))).))))))..(((((...((((((....))))))....-....))))).)..))) ( -35.50) >consensus UGGCAUUGGGGGA_AAGUGGUGAAAGGAGUUAGGCCUCGGUUCGACCAUAUUUCCUAUGGCCUGGGGCCCAUGGAAGUGGGCGGAA_AAGGGGGAUGCUGGUG .(((....(((((...((((((((..(((......)))..))).)))))..)))))...)))....((((((....))))))..................... (-32.49 = -32.27 + -0.22)

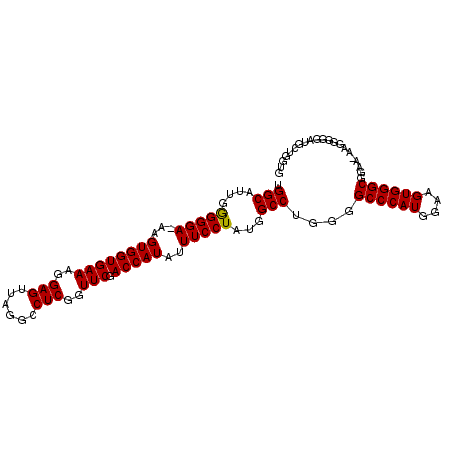

| Location | 13,567,575 – 13,567,672 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 89.90 |
| Mean single sequence MFE | -19.58 |
| Consensus MFE | -16.86 |
| Energy contribution | -17.10 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.86 |
| SVM decision value | 2.39 |
| SVM RNA-class probability | 0.993378 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13567575 97 + 23771897 CUUCCAUGGGCCCCAGGCCAUAGGAAAUAUGGUCGAACCGAGGCCUAACUCCUUUCACCACUUUUCCCCCAAUGCCA--------------CUUUCCACUUCCCAUACGAU .....(((((((...))))...(((((..((((.(((..(((......)))..)))))))..)))))..........--------------............)))..... ( -22.60) >DroSec_CAF1 49139 96 + 1 CUUCCAUGGGCCCCAGGCCAUAGGAAAUAUGGUCGAACCGAGGCCUAACUCCUUUCACCACUU-UCCCUCAAUGCCA--------------CUUUCCACUUUCCAUACGAU .....(((((((...))))...(((((..((((.(((..(((......)))..))))))).))-)))..........--------------............)))..... ( -20.00) >DroSim_CAF1 35302 96 + 1 CUUCCAUGGGCCCCAGGCCAUAGGAAAUAUGGUCGAACCGAGGCCUAACUCCUUUCACCACUU-UCCCCCAAUGCCA--------------CUUUCCACUUUCCAUACGAU .....(((((((...))))...(((((..((((.(((..(((......)))..))))))).))-)))..........--------------............)))..... ( -20.00) >DroEre_CAF1 49915 110 + 1 CUUCCAUCGGCCCCAGGCCAUAGGAAAUAUGGUCGAACCGAGGCCUAACUCCUUUUACCACUU-UCCCCCAAUGCCACUGUCCACUUUCCACUUUCCACUUUCCAUACGAU .....(((((((...))))...(((((..((((.(((..(((......)))..))))))).))-))).........................................))) ( -18.10) >DroYak_CAF1 51681 96 + 1 CUUCCAAAACCCUCGGGCCAUAGGAAAUAUGGUCGAACCGAGGCCUAACUCCUUUCACCACUU-UCCCCCAAUGCCA--------------CUUUCCACUUUCCAUGCGAU ...............(((....(((((..((((.(((..(((......)))..))))))).))-)))......))).--------------.................... ( -17.20) >consensus CUUCCAUGGGCCCCAGGCCAUAGGAAAUAUGGUCGAACCGAGGCCUAACUCCUUUCACCACUU_UCCCCCAAUGCCA______________CUUUCCACUUUCCAUACGAU .((((...((((...))))...))))...((((.(((..(((......)))..)))))))................................................... (-16.86 = -17.10 + 0.24)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:34:51 2006