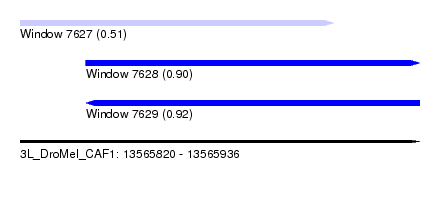
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,565,820 – 13,565,936 |
| Length | 116 |
| Max. P | 0.915593 |

| Location | 13,565,820 – 13,565,911 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 80.95 |
| Mean single sequence MFE | -26.82 |
| Consensus MFE | -18.82 |
| Energy contribution | -18.07 |
| Covariance contribution | -0.75 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.19 |
| Structure conservation index | 0.70 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.505619 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13565820 91 + 23771897 ---ACUAAGACAGACCUCGACC---UUACCUUUUGGGUGCAAUUUGCUGCGGCAGU------GGCAGUGGCAGCAAUUAAGCCAUAGGGUAGAUUAACAUUAA ---...................---(((((((...(((.....(((((((.((...------....)).)))))))....)))..)))))))........... ( -25.70) >DroPse_CAF1 55059 91 + 1 AGAGCUGAGACUGCUCUGCUUCUGUUGGCCUUUUGGGUACAAUUUGCAGUGGC------------AGUGGCAGCAAUUAAGCCAUGAGGUAGAUUAACUUGAA ...((((..((((((((((....(((((((.....))).))))..)))).)))------------)))..))))......(((....)))............. ( -30.60) >DroSec_CAF1 47392 91 + 1 ---ACUAAGACAGACCUCGACC---UUGCCUUUUGGGUGCAAUUUGCUGCGGCAGU------GGCAGUGGCAGUAAUUAAGCCACAGGGUAGAUUAACAUUAA ---.............((.(((---(((((.....(.((((......)))).)...------))).(((((.........)))))))))).)).......... ( -23.50) >DroSim_CAF1 33499 91 + 1 ---ACUAAGACAGACCUCGACC---UUACCUUUUGGGUGCAAUUUGCUGCGGCACU------GGCAGUGGCAGCAAUUAAGCCAUAGGGUAGAUUAACAUUAA ---...................---(((((((...(((.....(((((((.((...------....)).)))))))....)))..)))))))........... ( -25.70) >DroEre_CAF1 48126 91 + 1 ---ACUUAGACAGACCUCGACC---UUACCUUUUGGGUGCAAUUUGCGGCGGCAGU------GGCAGUGGCAGCAAUUUAGCCAUGGGGUAGAUUAACAUUAA ---..........(((((.(((---(........))))......(((.((....))------.)))(((((.........))))))))))............. ( -21.70) >DroPer_CAF1 54818 103 + 1 AGAACUGAGACUGCUCUGCUUCUGUUGGCCUUUUGGGUACAAUUUGCAGUGGCAGUGGCAGUGGCAGUGGCAGCAAUUAAGCCAUGAGGUAGAUUAACUUGAA ...((((..(((((((((((.((((..(((.....))).......)))).))))).))))))..))))(((.........))).(.((((......)))).). ( -33.70) >consensus ___ACUAAGACAGACCUCGACC___UUACCUUUUGGGUGCAAUUUGCUGCGGCAGU______GGCAGUGGCAGCAAUUAAGCCAUAGGGUAGAUUAACAUUAA .........................(((((((...(((.....((((.((.((.............)).)).))))....)))..)))))))........... (-18.82 = -18.07 + -0.75)


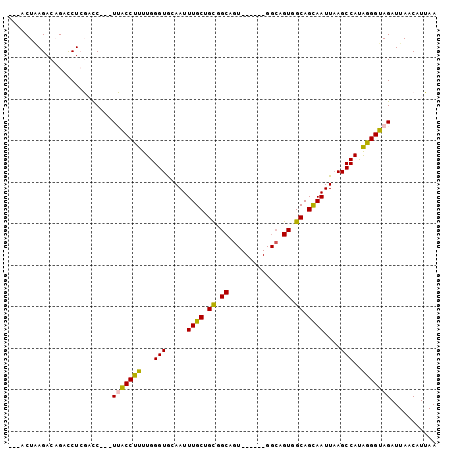
| Location | 13,565,839 – 13,565,936 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 80.69 |
| Mean single sequence MFE | -37.48 |
| Consensus MFE | -24.30 |
| Energy contribution | -25.35 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.65 |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.901541 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13565839 97 + 23771897 --UUACCUUUUGGGUGCAAUUUGCUGCGGCAGU------GGCAGUGGCAGCAAUUAAGCCAUAGGGUAGAUUAACAUUAAGCUGCCGCCA--------GAGCGCGAUGCACUU --...((....))(((((.(.((((((((((((------....(((((.........)))))...((......)).....))))))))..--------.)))).).))))).. ( -32.20) >DroPse_CAF1 55082 101 + 1 GUUGGCCUUUUGGGUACAAUUUGCAGUGGC------------AGUGGCAGCAAUUAAGCCAUGAGGUAGAUUAACUUGAAGCUGCCGCUGCCUUUGACGAGACAGAUGCGCUC (((((((.....))).))))((((((.(((------------(((((((((......(((....)))((.....))....)))))))))))).))).)))............. ( -34.10) >DroSec_CAF1 47411 96 + 1 --UUGCCUUUUGGGUGCAAUUUGCUGCGGCAGU------GGCAGUGGCAGUAAUUAAGCCACAGGGUAGAUUAACAUUAAGCUGCCGCCG--------GAGCGCGAUGCACU- --...((....))(((((.(.((((.((((...------....(((((.........)))))..(((((.(((....))).)))))))))--------.)))).).))))).- ( -37.60) >DroEre_CAF1 48145 97 + 1 --UUACCUUUUGGGUGCAAUUUGCGGCGGCAGU------GGCAGUGGCAGCAAUUUAGCCAUGGGGUAGAUUAACAUUAAGCUGCCGCCG--------GGGCAAGAUGCACUU --...((....))(((((.((((((((((((((------...((((.....((((((.((...)).))))))..))))..))))))))).--------..))))).))))).. ( -39.10) >DroYak_CAF1 49877 96 + 1 --UUACCUUUUGGGUGCAAUUUGCUGCGGCAGU------GGCAGUGGCAGCAAUUAAGCCACAGGGUAGAUUAACAUUAAGCUGCCGCCG--------GGGCAAGAUGCACU- --...((....))(((((.((((((.((((...------....(((((.........)))))..(((((.(((....))).)))))))))--------.)))))).))))).- ( -41.80) >DroPer_CAF1 54841 113 + 1 GUUGGCCUUUUGGGUACAAUUUGCAGUGGCAGUGGCAGUGGCAGUGGCAGCAAUUAAGCCAUGAGGUAGAUUAACUUGAAGCUGCCGCUGCCUUUGACGAGACAGAUGCGCUC (((((((.....))).))))((((((.((((((((((((..(.(((((.........))))).((((......)))))..)))))))))))).))).)))............. ( -40.10) >consensus __UUACCUUUUGGGUGCAAUUUGCUGCGGCAGU______GGCAGUGGCAGCAAUUAAGCCAUAGGGUAGAUUAACAUUAAGCUGCCGCCG________GAGCAAGAUGCACU_ .....((....))(((((.(((((.((((((((..........(((((.........)))))...((......)).....))))))))............))))).))))).. (-24.30 = -25.35 + 1.06)

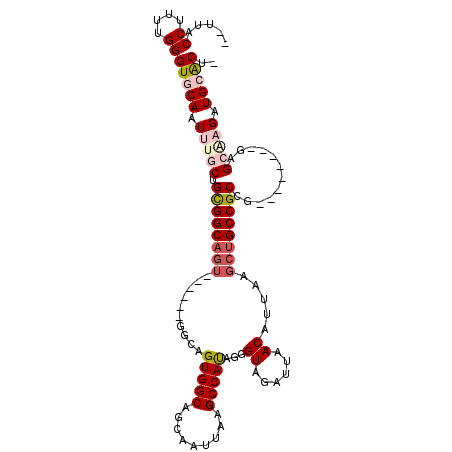
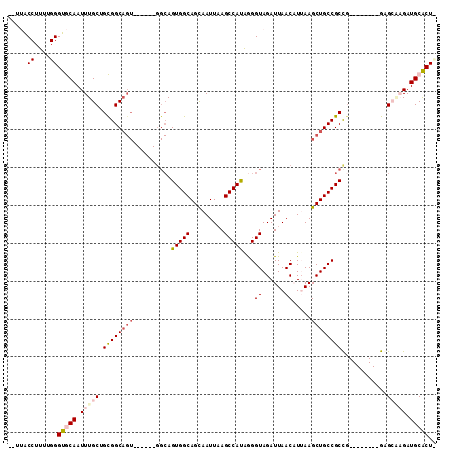
| Location | 13,565,839 – 13,565,936 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 80.69 |
| Mean single sequence MFE | -31.23 |
| Consensus MFE | -19.90 |
| Energy contribution | -20.35 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.59 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.915593 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13565839 97 - 23771897 AAGUGCAUCGCGCUC--------UGGCGGCAGCUUAAUGUUAAUCUACCCUAUGGCUUAAUUGCUGCCACUGCC------ACUGCCGCAGCAAAUUGCACCCAAAAGGUAA-- ..(((((..((((..--------((((((((((.....(((((.(((.....))).))))).))))))...)))------)..).))).......)))))((....))...-- ( -28.10) >DroPse_CAF1 55082 101 - 1 GAGCGCAUCUGUCUCGUCAAAGGCAGCGGCAGCUUCAAGUUAAUCUACCUCAUGGCUUAAUUGCUGCCACU------------GCCACUGCAAAUUGUACCCAAAAGGCCAAC (((.((....)))))(((...(((((.((((((...((((((..........))))))....)))))).))------------)))..(((.....))).......))).... ( -30.40) >DroSec_CAF1 47411 96 - 1 -AGUGCAUCGCGCUC--------CGGCGGCAGCUUAAUGUUAAUCUACCCUGUGGCUUAAUUACUGCCACUGCC------ACUGCCGCAGCAAAUUGCACCCAAAAGGCAA-- -.(((((....(((.--------((((((..((.....((......))...(((((.........))))).)).------.)))))).)))....)))))...........-- ( -30.70) >DroEre_CAF1 48145 97 - 1 AAGUGCAUCUUGCCC--------CGGCGGCAGCUUAAUGUUAAUCUACCCCAUGGCUAAAUUGCUGCCACUGCC------ACUGCCGCCGCAAAUUGCACCCAAAAGGUAA-- ..(((((..((((..--------.((((((((......((......))....((((.........)))).....------.))))))))))))..)))))((....))...-- ( -32.00) >DroYak_CAF1 49877 96 - 1 -AGUGCAUCUUGCCC--------CGGCGGCAGCUUAAUGUUAAUCUACCCUGUGGCUUAAUUGCUGCCACUGCC------ACUGCCGCAGCAAAUUGCACCCAAAAGGUAA-- -.(((((..((((..--------((((((((((.....(((((.((((...)))).))))).))))))......------...))))..))))..)))))((....))...-- ( -33.10) >DroPer_CAF1 54841 113 - 1 GAGCGCAUCUGUCUCGUCAAAGGCAGCGGCAGCUUCAAGUUAAUCUACCUCAUGGCUUAAUUGCUGCCACUGCCACUGCCACUGCCACUGCAAAUUGUACCCAAAAGGCCAAC (((.((....)))))......(((((.((((((...((((((..........))))))....)))))).)))))...(((..(((....)))..(((....)))..))).... ( -33.10) >consensus _AGUGCAUCUCGCUC________CGGCGGCAGCUUAAUGUUAAUCUACCCCAUGGCUUAAUUGCUGCCACUGCC______ACUGCCGCAGCAAAUUGCACCCAAAAGGCAA__ ..(((((................(((.((((((.....(((((.(((.....))).))))).)))))).)))..........(((....)))...)))))............. (-19.90 = -20.35 + 0.45)

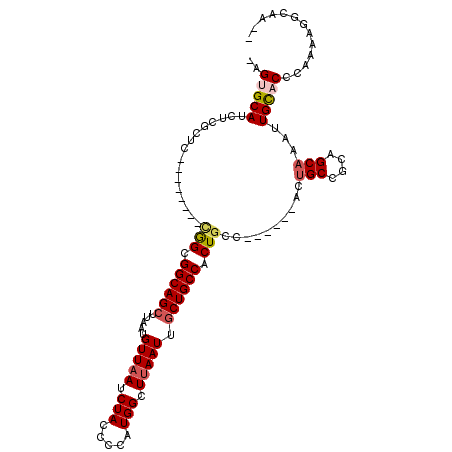
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:34:48 2006