
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,563,438 – 13,563,637 |
| Length | 199 |
| Max. P | 0.997072 |

| Location | 13,563,438 – 13,563,532 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 78.79 |
| Mean single sequence MFE | -21.01 |
| Consensus MFE | -9.67 |
| Energy contribution | -9.39 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.46 |
| SVM decision value | 1.14 |
| SVM RNA-class probability | 0.921363 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13563438 94 + 23771897 UGCGAGUGUAUGUGUGCGUGUGCGUUUGACACACAAUAAACACCAAUUAAGAAAAUAAUAUUUUGUGUACACACAC------AAGCAAACA-AAUGUCU-------CU (((...(((.((((((((((((.......)))))....................((((....))))))))))).))------).)))....-.......-------.. ( -21.50) >DroSec_CAF1 45069 86 + 1 UG--------------CGUGUGCGUUUGACACACAAUAAACACCAAUUAAGAAAAUAAUAUUUUGUGUACACACACACACAGAAGCAAACA-AAUGUCU-------CU ..--------------.(((((.((.(((((((.((((....................)))).))))).)).)).)))))(((..((....-..))..)-------)) ( -17.95) >DroSim_CAF1 31166 98 + 1 UGCG--UGUGGGUGUGCGUGUGCGUUUGACACACAAUAAACACCAAUUAAGAAAAUAAUAUUUUGUGUACACACAUACACAGAAGCAAACA-AAUGUCU-------CU (((.--....(((((...((((.(.....).))))....)))))................(((((((((......))))))))))))....-.......-------.. ( -23.80) >DroYak_CAF1 47459 89 + 1 ---G--UGUGUGUGUGGGUGUGCGUUUGACACACAAUAAACACCAAUUAAGAAAAUAAUAUUUUGUGUACACACAC------AGGCAAACA-AAUGUCU-------CU ---.--(((((((((((((((....(((.....)))...)))))..........((((....)))).)))))))))------)((((....-..)))).-------.. ( -24.00) >DroMoj_CAF1 62522 86 + 1 U------------GUGUGUGUGUGUGUGACACACAAUAAACACCAAUUAAGAAUAUAAUAUUUUGUGUAUACAGGC------A----AACAAAAUGUCUCCCCUGGCU .------------((((.((((((.....))))))....)))).........((((((....))))))...((((.------.----.((.....))....))))... ( -17.80) >consensus UG_G__UGU__GUGUGCGUGUGCGUUUGACACACAAUAAACACCAAUUAAGAAAAUAAUAUUUUGUGUACACACAC______AAGCAAACA_AAUGUCU_______CU .................(((((.((...(((((.((((....................)))).))))))).)))))................................ ( -9.67 = -9.39 + -0.28)



| Location | 13,563,438 – 13,563,532 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 78.79 |
| Mean single sequence MFE | -24.36 |
| Consensus MFE | -14.16 |
| Energy contribution | -15.00 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.98 |
| Structure conservation index | 0.58 |
| SVM decision value | 2.79 |
| SVM RNA-class probability | 0.997072 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13563438 94 - 23771897 AG-------AGACAUU-UGUUUGCUU------GUGUGUGUACACAAAAUAUUAUUUUCUUAAUUGGUGUUUAUUGUGUGUCAAACGCACACGCACACAUACACUCGCA .(-------((.....-(((.(((.(------((((((.((((((((((((((.((....)).)))))))..)))))))....))))))).))).)))....)))... ( -27.20) >DroSec_CAF1 45069 86 - 1 AG-------AGACAUU-UGUUUGCUUCUGUGUGUGUGUGUACACAAAAUAUUAUUUUCUUAAUUGGUGUUUAUUGUGUGUCAAACGCACACG--------------CA ((-------(..((..-....))..)))((((((((((.((((((((((((((.((....)).)))))))..)))))))....)))))))))--------------). ( -26.60) >DroSim_CAF1 31166 98 - 1 AG-------AGACAUU-UGUUUGCUUCUGUGUAUGUGUGUACACAAAAUAUUAUUUUCUUAAUUGGUGUUUAUUGUGUGUCAAACGCACACGCACACCCACA--CGCA ..-------((((...-.))))((...((((..(((((((((((.....((((......))))..)))).....(((((.....))))))))))))..))))--.)). ( -25.70) >DroYak_CAF1 47459 89 - 1 AG-------AGACAUU-UGUUUGCCU------GUGUGUGUACACAAAAUAUUAUUUUCUUAAUUGGUGUUUAUUGUGUGUCAAACGCACACCCACACACACA--C--- ..-------((((...-.))))...(------((((((((((((.....((((......))))..))))....((((((.....))))))...)))))))))--.--- ( -23.60) >DroMoj_CAF1 62522 86 - 1 AGCCAGGGGAGACAUUUUGUU----U------GCCUGUAUACACAAAAUAUUAUAUUCUUAAUUGGUGUUUAUUGUGUGUCACACACACACACAC------------A .....(((.((((.....)))----)------.)))(((.((((.....((((......))))..)))).)))(((((((.....)))))))...------------. ( -18.70) >consensus AG_______AGACAUU_UGUUUGCUU______GUGUGUGUACACAAAAUAUUAUUUUCUUAAUUGGUGUUUAUUGUGUGUCAAACGCACACGCACAC__ACA__C_CA .........((((.....))))..........(((((((((((((((((((((..........))))))))....)))))...))))))))................. (-14.16 = -15.00 + 0.84)

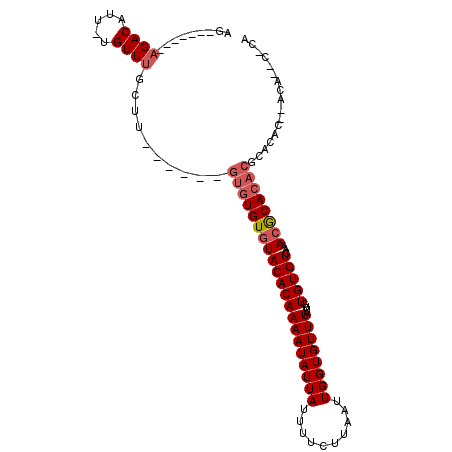

| Location | 13,563,532 – 13,563,637 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 77.68 |
| Mean single sequence MFE | -23.43 |
| Consensus MFE | -12.04 |
| Energy contribution | -12.15 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.51 |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.901167 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13563532 105 + 23771897 GGCAAUUUGAAAAGCCAGCGCCA------AAGCGUU--CGUCUCUAG---UUU-UGUACAAGUGCUGUGCAAAUAAUGCACAAAAGUAUUCUACAAA-AUGCGAACAACAAACAACCG (((..........))).((....------..))(((--(((.....(---(((-((((..(((((((((((.....)))))...)))))).))))))-))))))))............ ( -27.30) >DroVir_CAF1 66186 94 + 1 GGCAAAUUGAAAAGCCAAGUC-------------UU--UCGCUCUGGACUUUU-CCUGCAAGUGCUGUGCAAAUAAUGCACAAAAGUAUGCUACAAAAAUACAAACAA-------AC- ((((.(((((((((((((((.-------------..--..))).))).)))))-)..((....))((((((.....))))))..))).))))................-------..- ( -20.60) >DroPse_CAF1 52394 99 + 1 GGCAAAUUGAAAAGCCAAAGCCAAG--CGACGCCUU--CGUCUCUAG---UUC-CGUACAAGUGCUGUGCAAAUAAUGCACAAAAGUAUGCUACAAA-AUACGAACAA---------- (((...(((......))).))).((--.((((....--)))).)).(---(((-.(((....)))((((((.....))))))...((((........-))))))))..---------- ( -22.30) >DroGri_CAF1 63264 103 + 1 GGCAAAUUGAAAAGCCAAGUC-A------AGAC-UUGCUCUCACUGGACUUUUCUCUGCAAGUGCUGUGCAAAUAAUGCACAAAAGUAUGCUACAAAAAUACAAACAA--A----AC- .(((....(((((((((.((.-.------(((.-....))).))))).))))))..)))......((((((.....))))))...((((.........))))......--.----..- ( -20.80) >DroYak_CAF1 47548 111 + 1 GGCAAUUUGAAAAGCCAACGCCAAAGCCAAAGCGUU--CGUCUCUAG---UUU-UGUACAAGUGCUGUGCAAAUAGUGCACAAAAGUAUGCUACAAA-AUGCGAACAACAAACAACCG ((...((((....((....((....))....))(((--(((.....(---(((-((((...((((((((((.....)))))...)))))..))))))-))))))))..))))...)). ( -27.30) >DroPer_CAF1 52244 99 + 1 GGCAAAUUGAAAAGCCAAAGCCAAG--CGACGCCUU--CGUCUCUAG---UUC-CGUACAAGUGCUGUGCAAAUAAUGCACAAAAGUAUGCUACAAA-AUACGAACAA---------- (((...(((......))).))).((--.((((....--)))).)).(---(((-.(((....)))((((((.....))))))...((((........-))))))))..---------- ( -22.30) >consensus GGCAAAUUGAAAAGCCAAAGCCA______AAGC_UU__CGUCUCUAG___UUU_CGUACAAGUGCUGUGCAAAUAAUGCACAAAAGUAUGCUACAAA_AUACGAACAA________C_ (((..........))).......................................(((...((((((((((.....)))))...)))))..)))........................ (-12.04 = -12.15 + 0.11)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:34:43 2006