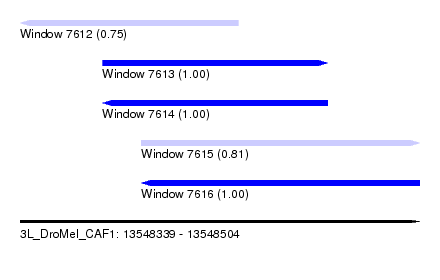
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,548,339 – 13,548,504 |
| Length | 165 |
| Max. P | 0.999562 |

| Location | 13,548,339 – 13,548,429 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 87.85 |
| Mean single sequence MFE | -29.02 |
| Consensus MFE | -21.76 |
| Energy contribution | -22.96 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.750693 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13548339 90 - 23771897 UAAAAAAAAUGGCGCCCAACGUGGGGCAAAGCUGAGGCUGCUGUUUUCUAUAUGAA-------GGUAUGGGAGAACCGGGAGUUUCUUCUUAACGCU ..........(((((((......)))).(((..((((((.(((((((((((((...-------.))))))))))..))).))))))..)))...))) ( -29.40) >DroSec_CAF1 35520 90 - 1 UAAAAAAAAUGGCGCCCAACGUGGGACAGAGCUGAGGCUGCUGUUUUCUAUAUGAA-------GGUAUGAGAGAACCGGCAGUUUCUUCUUAACGCU ..........(((((((.....)))...(((..((((((((((((((((((((...-------.)))).)))))).))))))))))..)))..)))) ( -30.40) >DroSim_CAF1 21618 89 - 1 UAAAA-AAAUGGCGCCCAACAUGGGGCAAAGCUGAGGCUGCUGUUUUCUAUAUGAA-------GGUGUGGGAGAACCGGGAGUUUCUUCUUAACGCU .....-....(((((((......)))).(((..((((((.(((((((((((((...-------.))))))))))..))).))))))..)))...))) ( -29.40) >DroEre_CAF1 36079 88 - 1 CAAAA-AAAUGGCGCCCAACGUGGGGCAGAGCUGAGGCUGCUGUUUUCUAUAUG-------GGGGAAUGGG-GCACCGGCAGUUUCUUCUUAACGCU .....-....(((((((......)))).(((..((((((((((((..((.....-------))..)))((.-...)))))))))))..)))...))) ( -30.90) >DroYak_CAF1 37566 94 - 1 CGAA--AAAUGGCGCCCAACGUGGGGCAGAGCUGAGGCUGCUGUUUUCUAUAUGAAGGUACGGGGUAUGGG-GUAACGAGAGUUUCUUCUUAACGCU .(((--((.((((((((......))))..(((....)))))))))))).............((.((..(((-(.(((....))).))))...)).)) ( -25.00) >consensus UAAAA_AAAUGGCGCCCAACGUGGGGCAGAGCUGAGGCUGCUGUUUUCUAUAUGAA_______GGUAUGGGAGAACCGGGAGUUUCUUCUUAACGCU ..........(((((((......)))).(((..((((((.(((((((((((((...........))))))))))..))).))))))..)))...))) (-21.76 = -22.96 + 1.20)



| Location | 13,548,373 – 13,548,466 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 90.12 |
| Mean single sequence MFE | -28.96 |
| Consensus MFE | -23.92 |
| Energy contribution | -24.04 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.96 |
| Structure conservation index | 0.83 |
| SVM decision value | 3.14 |
| SVM RNA-class probability | 0.998565 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13548373 93 + 23771897 ----UUCAUAUAGAAAACAGCAGCCUCAGCUUUGCCCCACGUUGGGCGCCAUUUUUUUUAUACGGCACACCA--GUGCCCAACGUUGGGCGCCAUUUUU ----........(((((..(((((....)))..((((.((((((((((((.............)))((....--))))))))))).))))))..))))) ( -33.22) >DroSec_CAF1 35554 93 + 1 ----UUCAUAUAGAAAACAGCAGCCUCAGCUCUGUCCCACGUUGGGCGCCAUUUUUUUUAUACGGCACACCA--UUGCCCCACCUUGGGCGCCAUUUUU ----............((((.(((....))))))).........((((((.............((((.....--.)))).(.....)))))))...... ( -22.90) >DroSim_CAF1 21652 92 + 1 ----UUCAUAUAGAAAACAGCAGCCUCAGCUUUGCCCCAUGUUGGGCGCCAUUU-UUUUAUACGGCACACCA--UUGCCGCACGUUGGGCGCCAUUUUU ----........(((((..((.(((.((((...((((......)))).......-.......(((((.....--.)))))...)))))))))..))))) ( -26.40) >DroEre_CAF1 36114 92 + 1 ------CAUAUAGAAAACAGCAGCCUCAGCUCUGCCCCACGUUGGGCGCCAUUU-UUUUGUAUGGCACACCACGUUGCCCCACGUUGGGCGCCAUUUUU ------......(((((..(((((....)))..((((.((((.(((((((((..-......)))))((.....)).)))).)))).))))))..))))) ( -32.20) >DroYak_CAF1 37602 95 + 1 UACCUUCAUAUAGAAAACAGCAGCCUCAGCUCUGCCCCACGUUGGGCGCCAUUU--UUCGUAUGGCUCACCA--GUGCCCCACGUUGGGCGCCAUUUUU ............(((((..(((((....)))..((((.((((.(((((((((..--.....)))))......--..)))).)))).))))))..))))) ( -30.10) >consensus ____UUCAUAUAGAAAACAGCAGCCUCAGCUCUGCCCCACGUUGGGCGCCAUUU_UUUUAUACGGCACACCA__UUGCCCCACGUUGGGCGCCAUUUUU ............(((((..(((((....)))..((((.((((.(((((((.............)))..........)))).)))).))))))..))))) (-23.92 = -24.04 + 0.12)



| Location | 13,548,373 – 13,548,466 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 90.12 |
| Mean single sequence MFE | -35.56 |
| Consensus MFE | -29.50 |
| Energy contribution | -30.06 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.59 |
| Structure conservation index | 0.83 |
| SVM decision value | 3.73 |
| SVM RNA-class probability | 0.999562 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13548373 93 - 23771897 AAAAAUGGCGCCCAACGUUGGGCAC--UGGUGUGCCGUAUAAAAAAAAUGGCGCCCAACGUGGGGCAAAGCUGAGGCUGCUGUUUUCUAUAUGAA---- .((((((((((((.(((((((((((--....))(((((.........)))))))))))))).))))..(((....))))))))))).........---- ( -41.00) >DroSec_CAF1 35554 93 - 1 AAAAAUGGCGCCCAAGGUGGGGCAA--UGGUGUGCCGUAUAAAAAAAAUGGCGCCCAACGUGGGACAGAGCUGAGGCUGCUGUUUUCUAUAUGAA---- .........((((......))))..--(((.(((((((.........))))))))))..((((((((((((....))).)))))..)))).....---- ( -30.70) >DroSim_CAF1 21652 92 - 1 AAAAAUGGCGCCCAACGUGCGGCAA--UGGUGUGCCGUAUAAAA-AAAUGGCGCCCAACAUGGGGCAAAGCUGAGGCUGCUGUUUUCUAUAUGAA---- .(((((((((((((.(((((((((.--.....))))))))....-.......((((......))))...).)).))).)))))))).........---- ( -31.80) >DroEre_CAF1 36114 92 - 1 AAAAAUGGCGCCCAACGUGGGGCAACGUGGUGUGCCAUACAAAA-AAAUGGCGCCCAACGUGGGGCAGAGCUGAGGCUGCUGUUUUCUAUAUG------ .((((((((((((.((((.((((.((.....))(((((......-..))))))))).)))).))))..(((....))))))))))).......------ ( -37.30) >DroYak_CAF1 37602 95 - 1 AAAAAUGGCGCCCAACGUGGGGCAC--UGGUGAGCCAUACGAA--AAAUGGCGCCCAACGUGGGGCAGAGCUGAGGCUGCUGUUUUCUAUAUGAAGGUA .((((((((((((.((((.((((.(--....).(((((.....--..))))))))).)))).))))..(((....)))))))))))............. ( -37.00) >consensus AAAAAUGGCGCCCAACGUGGGGCAA__UGGUGUGCCGUAUAAAA_AAAUGGCGCCCAACGUGGGGCAGAGCUGAGGCUGCUGUUUUCUAUAUGAA____ .((((((((((((.((((.((((..........(((((.........))))))))).)))).))))..(((....)))))))))))............. (-29.50 = -30.06 + 0.56)

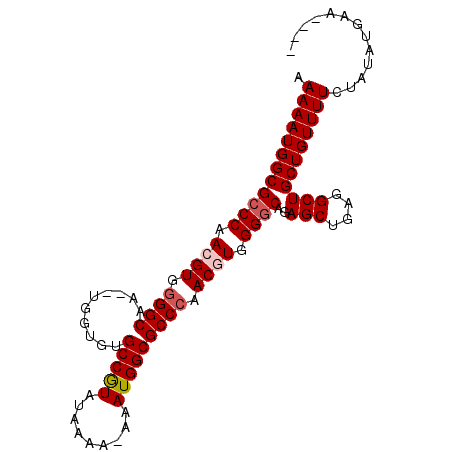
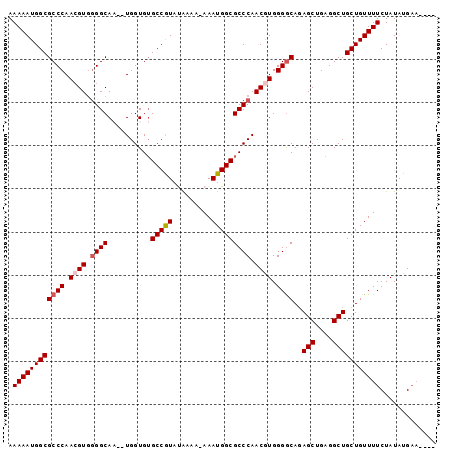
| Location | 13,548,389 – 13,548,504 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 93.94 |
| Mean single sequence MFE | -36.05 |
| Consensus MFE | -31.12 |
| Energy contribution | -31.24 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.810944 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13548389 115 + 23771897 CAGCCUCAGCUUUGCCCCACGUUGGGCGCCAUUUUUUUUAUACGGCACACCA--GUGCCCAACGUUGGGCGCCAUUUUUGUCAAGUCGCAUUAAGGCAGCGCGCUCAUUCGACUUGU .(((....)))..((((.((((((((((((.............)))((....--))))))))))).))))...........(((((((.((...(((.....))).)).))))))). ( -40.42) >DroSec_CAF1 35570 115 + 1 CAGCCUCAGCUCUGUCCCACGUUGGGCGCCAUUUUUUUUAUACGGCACACCA--UUGCCCCACCUUGGGCGCCAUUUUUGUCAAGUCGCAUUAAGGCAGCGCGCUCAUUCGACUUGU ..(((.((((..........)))))))(((.............)))......--..((((......))))...........(((((((.((...(((.....))).)).))))))). ( -28.82) >DroSim_CAF1 21668 114 + 1 CAGCCUCAGCUUUGCCCCAUGUUGGGCGCCAUUU-UUUUAUACGGCACACCA--UUGCCGCACGUUGGGCGCCAUUUUUGUCAAGUCGCAUUAAGGCAGCGCGCUCAUUCGACUUGU ..(((.((((...((((......)))).......-.......(((((.....--.)))))...)))))))...........(((((((.((...(((.....))).)).))))))). ( -34.30) >DroEre_CAF1 36128 116 + 1 CAGCCUCAGCUCUGCCCCACGUUGGGCGCCAUUU-UUUUGUAUGGCACACCACGUUGCCCCACGUUGGGCGCCAUUUUUGUCAAGUCGCAUUAAGGCAGCGCGCUCAUUCGACUUGU .(((....)))..((((.((((.(((((((((..-......)))))((.....)).)))).)))).))))...........(((((((.((...(((.....))).)).))))))). ( -39.40) >DroYak_CAF1 37622 113 + 1 CAGCCUCAGCUCUGCCCCACGUUGGGCGCCAUUU--UUCGUAUGGCUCACCA--GUGCCCCACGUUGGGCGCCAUUUUUGUCAAGUCGCAUUAAGGCAGCGCGCUCAUUCGACUUGU .(((....)))..((((.((((.(((((((((..--.....)))))......--..)))).)))).))))...........(((((((.((...(((.....))).)).))))))). ( -37.30) >consensus CAGCCUCAGCUCUGCCCCACGUUGGGCGCCAUUU_UUUUAUACGGCACACCA__UUGCCCCACGUUGGGCGCCAUUUUUGUCAAGUCGCAUUAAGGCAGCGCGCUCAUUCGACUUGU .(((....)))..((((.((((.(((((((.............)))..........)))).)))).))))...........(((((((.((...(((.....))).)).))))))). (-31.12 = -31.24 + 0.12)

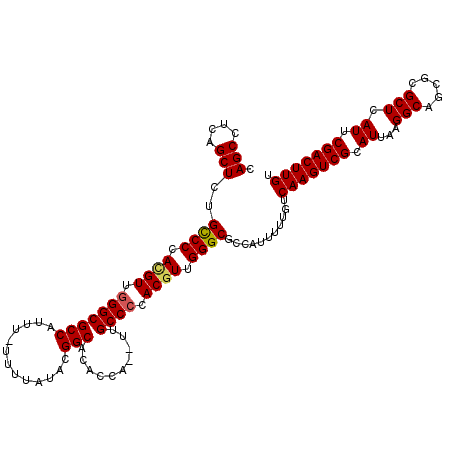
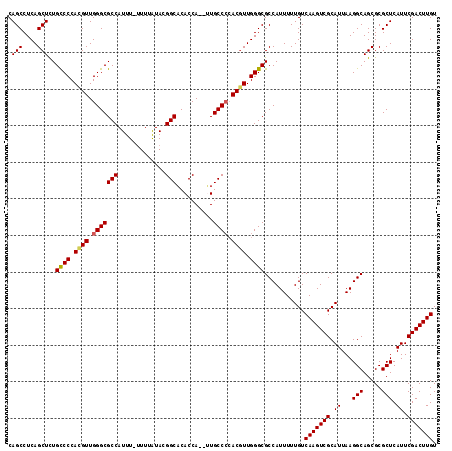
| Location | 13,548,389 – 13,548,504 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 93.94 |
| Mean single sequence MFE | -42.62 |
| Consensus MFE | -36.60 |
| Energy contribution | -37.16 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.86 |
| SVM decision value | 3.08 |
| SVM RNA-class probability | 0.998358 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13548389 115 - 23771897 ACAAGUCGAAUGAGCGCGCUGCCUUAAUGCGACUUGACAAAAAUGGCGCCCAACGUUGGGCAC--UGGUGUGCCGUAUAAAAAAAAUGGCGCCCAACGUGGGGCAAAGCUGAGGCUG .(((((((..((((.((...))))))...)))))))...........((((.(((((((((((--....))(((((.........)))))))))))))).))))..(((....))). ( -48.10) >DroSec_CAF1 35570 115 - 1 ACAAGUCGAAUGAGCGCGCUGCCUUAAUGCGACUUGACAAAAAUGGCGCCCAAGGUGGGGCAA--UGGUGUGCCGUAUAAAAAAAAUGGCGCCCAACGUGGGACAGAGCUGAGGCUG .(((((((..((((.((...))))))...)))))))........(((((((......))))..--(((.(((((((.........))))))))))............)))....... ( -37.60) >DroSim_CAF1 21668 114 - 1 ACAAGUCGAAUGAGCGCGCUGCCUUAAUGCGACUUGACAAAAAUGGCGCCCAACGUGCGGCAA--UGGUGUGCCGUAUAAAA-AAAUGGCGCCCAACAUGGGGCAAAGCUGAGGCUG .(((((((..((((.((...))))))...)))))))........((((((....((((((((.--.....))))))))....-....)))((((......))))...)))....... ( -38.90) >DroEre_CAF1 36128 116 - 1 ACAAGUCGAAUGAGCGCGCUGCCUUAAUGCGACUUGACAAAAAUGGCGCCCAACGUGGGGCAACGUGGUGUGCCAUACAAAA-AAAUGGCGCCCAACGUGGGGCAGAGCUGAGGCUG .(((((((..((((.((...))))))...)))))))...........((((.((((.((((.((.....))(((((......-..))))))))).)))).))))..(((....))). ( -44.40) >DroYak_CAF1 37622 113 - 1 ACAAGUCGAAUGAGCGCGCUGCCUUAAUGCGACUUGACAAAAAUGGCGCCCAACGUGGGGCAC--UGGUGAGCCAUACGAA--AAAUGGCGCCCAACGUGGGGCAGAGCUGAGGCUG .(((((((..((((.((...))))))...)))))))...........((((.((((.((((.(--....).(((((.....--..))))))))).)))).))))..(((....))). ( -44.10) >consensus ACAAGUCGAAUGAGCGCGCUGCCUUAAUGCGACUUGACAAAAAUGGCGCCCAACGUGGGGCAA__UGGUGUGCCGUAUAAAA_AAAUGGCGCCCAACGUGGGGCAGAGCUGAGGCUG .(((((((..((((.((...))))))...)))))))...........((((.((((.((((..........(((((.........))))))))).)))).))))..(((....))). (-36.60 = -37.16 + 0.56)

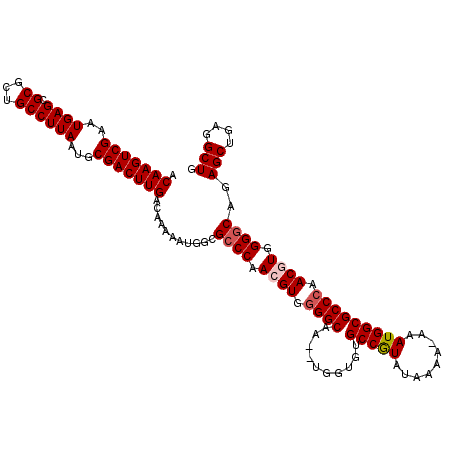

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:34:36 2006