
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,518,022 – 13,518,168 |
| Length | 146 |
| Max. P | 0.991218 |

| Location | 13,518,022 – 13,518,113 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.55 |
| Mean single sequence MFE | -42.17 |
| Consensus MFE | -21.99 |
| Energy contribution | -23.24 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.895110 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13518022 91 + 23771897 CUAACCAGAGAAGG--UC---CUUGGUCCUUGC-------UGCCUGCCGCCUGGUGCU-UGCCGC----------------UUGAGCCUGGACUCGUGCCAGGCGGCACUUUUGGCAGCA ..........((((--.(---....).))))((-------((((((((((((((..(.-.(((((----------------....))..)).)..)..)))))))))).....)))))). ( -45.80) >DroPse_CAF1 5805 109 + 1 CUAACAAGAGAGGUCUUC---CCCGGUGCCUGG-------UGGCUGGUGCCUGGUGCU-UGGUGCCUGGAUGCCUGGGUCCUUGUUCCUGGACUCGUGCCAGGCGGCACUUUUGGCAGCA ((..((((((.((.....---.))(.(((((((-------((((.((..((.......-.))..)).....))).((((((........))))))..)))))))).).))))))..)).. ( -46.90) >DroSec_CAF1 5367 84 + 1 CUAACCAGAGAAGG--UC---CUUCGUCCUUGC-------UGC-------CUGGUGCU-UGCCGC----------------UUGGGCCUGGACUCGUGCCAGGCGGCACUUUUGGCAGCA ..........((((--.(---....).))))((-------(((-------(.((((((-..((..----------------..))((((((.(....)))))))))))))...)))))). ( -36.30) >DroEre_CAF1 5339 91 + 1 CUAACCAGAGAAGG--UC---CUUGGUCCUUGC-------UGCCUGCUGCCUGGUGCU-UGCCGC----------------UUGGGCCUGGACUCGUGCCAGGCGGCACUUUUGGCAGCA ..........((((--.(---....).))))((-------((((((((((((((..(.-.(((((----------------....))..)).)..)..)))))))))).....)))))). ( -43.70) >DroYak_CAF1 5340 99 + 1 CUAACCAGAGAAGG--UC---CUUGGUCCUUGCCGCCUGCUGCCUGCUGCCUGGUGCUUUGCCGC----------------UUGGGCCUGGACUCGUGCCAGGCGGCACUUUUGGCAGCA .......(..((((--.(---....).))))..)....((((((((((((((((..(....((((----------------....))..))....)..)))))))))).....)))))). ( -43.50) >DroAna_CAF1 5625 86 + 1 CUAACCGGAGAGGC--UCUGGCCGGGUCCUUAG-------U-CUUGCUGCCUGGUGC------------------------UUGGUCCUGGACGCGUGCCAGGCGGCACUUUUGGCAGCA ((((..(((..(((--....)))...)))))))-------.-..(((((((.(((((------------------------(..(.(((((.......))))))))))))...))))))) ( -36.80) >consensus CUAACCAGAGAAGG__UC___CUUGGUCCUUGC_______UGCCUGCUGCCUGGUGCU_UGCCGC________________UUGGGCCUGGACUCGUGCCAGGCGGCACUUUUGGCAGCA ..........((((.............))))..............((((((.((((((...........................((((((.(....)))))))))))))...)))))). (-21.99 = -23.24 + 1.25)


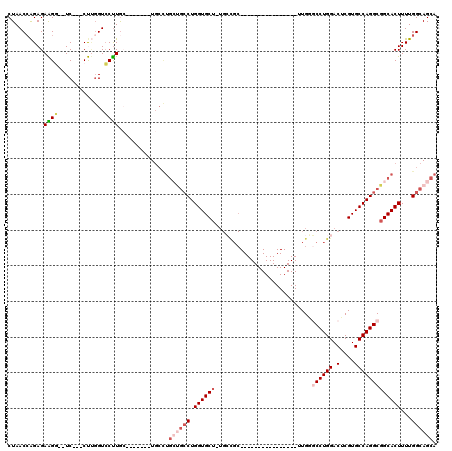
| Location | 13,518,022 – 13,518,113 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.55 |
| Mean single sequence MFE | -38.18 |
| Consensus MFE | -18.06 |
| Energy contribution | -19.78 |
| Covariance contribution | 1.72 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.47 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.827796 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13518022 91 - 23771897 UGCUGCCAAAAGUGCCGCCUGGCACGAGUCCAGGCUCAA----------------GCGGCA-AGCACCAGGCGGCAGGCA-------GCAAGGACCAAG---GA--CCUUCUCUGGUUAG (((((((.....((((((((((...((((....))))..----------------((....-.)).))))))))))))))-------)))..(((((..---(.--....)..))))).. ( -47.70) >DroPse_CAF1 5805 109 - 1 UGCUGCCAAAAGUGCCGCCUGGCACGAGUCCAGGAACAAGGACCCAGGCAUCCAGGCACCA-AGCACCAGGCACCAGCCA-------CCAGGCACCGGG---GAAGACCUCUCUUGUUAG .(.((((....(((((((((((.....((((........)))))))))).....)))))..-.......(((....))).-------...)))).)(((---(....))))......... ( -37.10) >DroSec_CAF1 5367 84 - 1 UGCUGCCAAAAGUGCCGCCUGGCACGAGUCCAGGCCCAA----------------GCGGCA-AGCACCAG-------GCA-------GCAAGGACGAAG---GA--CCUUCUCUGGUUAG (((((((....((((.(((((((....).))))))((..----------------..))..-.))))..)-------)))-------))).........---((--((......)))).. ( -32.50) >DroEre_CAF1 5339 91 - 1 UGCUGCCAAAAGUGCCGCCUGGCACGAGUCCAGGCCCAA----------------GCGGCA-AGCACCAGGCAGCAGGCA-------GCAAGGACCAAG---GA--CCUUCUCUGGUUAG (((((((.....(((((((((((....).))))))....----------------((....-.))....))))...))))-------)))..(((((..---(.--....)..))))).. ( -38.30) >DroYak_CAF1 5340 99 - 1 UGCUGCCAAAAGUGCCGCCUGGCACGAGUCCAGGCCCAA----------------GCGGCAAAGCACCAGGCAGCAGGCAGCAGGCGGCAAGGACCAAG---GA--CCUUCUCUGGUUAG (((((((.....(((((((((((....).))))))....----------------((......))....))))((.....)).)))))))..(((((..---(.--....)..))))).. ( -40.80) >DroAna_CAF1 5625 86 - 1 UGCUGCCAAAAGUGCCGCCUGGCACGCGUCCAGGACCAA------------------------GCACCAGGCAGCAAG-A-------CUAAGGACCCGGCCAGA--GCCUCUCCGGUUAG (((((((....((((..((((((....).))))).....------------------------))))..))))))).(-(-------((..(((...(((....--)))..))))))).. ( -32.70) >consensus UGCUGCCAAAAGUGCCGCCUGGCACGAGUCCAGGCCCAA________________GCGGCA_AGCACCAGGCAGCAGGCA_______GCAAGGACCAAG___GA__CCUUCUCUGGUUAG (((((((.........(((((((....).))))))....................((......))....)))))))............................................ (-18.06 = -19.78 + 1.72)

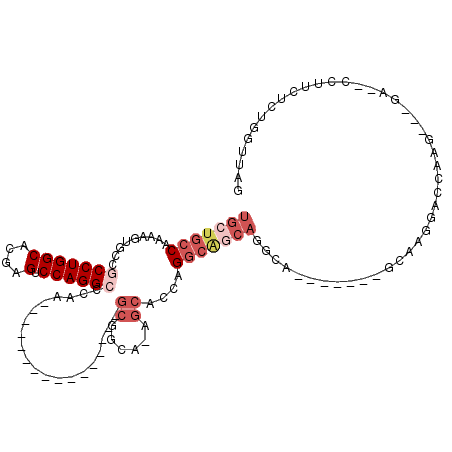
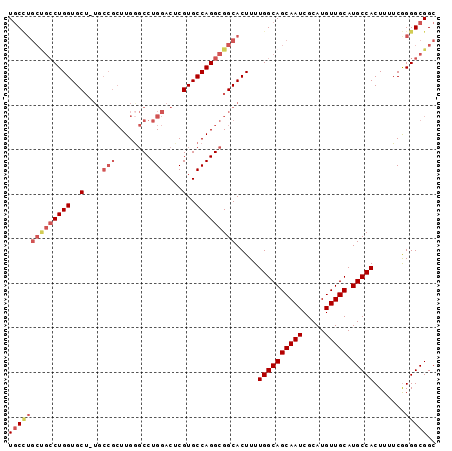
| Location | 13,518,050 – 13,518,145 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 90.99 |
| Mean single sequence MFE | -42.38 |
| Consensus MFE | -35.00 |
| Energy contribution | -36.96 |
| Covariance contribution | 1.96 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.83 |
| SVM decision value | 2.22 |
| SVM RNA-class probability | 0.990481 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13518050 95 + 23771897 UGCCUGCCGCCUGGUGCU-UGCCGCUUGAGCCUGGACUCGUGCCAGGCGGCACUUUUGGCAGCAAUCGCAUGUUGCAUGCCACUUUUCGGGGCGGC .(((((((((((((..(.-.(((((....))..)).)..)..))))))))).....((((((((((.....))))).))))).......))))... ( -47.10) >DroSec_CAF1 5395 88 + 1 UGC-------CUGGUGCU-UGCCGCUUGGGCCUGGACUCGUGCCAGGCGGCACUUUUGGCAGCAAUCGCAUGUUGCAUGCCACUUUUCGGGGCGGC .((-------((.(....-((((((((((((........)).))))))))))....((((((((((.....))))).))))).....).))))... ( -37.60) >DroEre_CAF1 5367 95 + 1 UGCCUGCUGCCUGGUGCU-UGCCGCUUGGGCCUGGACUCGUGCCAGGCGGCACUUUUGGCAGCAAUCGCAUGUUGCAUGCCACUUUUCGGGGCGGC .(((((((((((((..(.-.(((((....))..)).)..)..))))))))).....((((((((((.....))))).))))).......))))... ( -45.00) >DroYak_CAF1 5375 96 + 1 UGCCUGCUGCCUGGUGCUUUGCCGCUUGGGCCUGGACUCGUGCCAGGCGGCACUUUUGGCAGCAAUCGCAUGUUGCAUGCCACUUUUCGGGGCGGC .(((((((((((((..(....((((....))..))....)..))))))))).....((((((((((.....))))).))))).......))))... ( -44.70) >DroAna_CAF1 5656 87 + 1 U-CUUGCUGCCUGGUGC--------UUGGUCCUGGACGCGUGCCAGGCGGCACUUUUGGCAGCAAUCGCAUGUUGCAUGCCACUUUUCAGGGCGGC .-..((((((((((..(--------.((........)).)..))))))))))......((((((......)))))).((((.((....)))))).. ( -37.50) >consensus UGCCUGCUGCCUGGUGCU_UGCCGCUUGGGCCUGGACUCGUGCCAGGCGGCACUUUUGGCAGCAAUCGCAUGUUGCAUGCCACUUUUCGGGGCGGC ((((((((((((((..(....(((........)))....)..))))))))).....((((((((((.....))))).))))).......))))).. (-35.00 = -36.96 + 1.96)


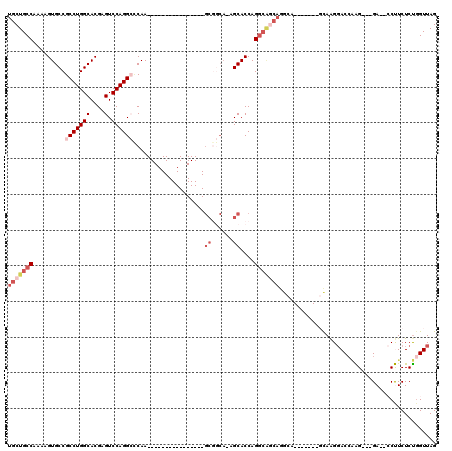
| Location | 13,518,050 – 13,518,145 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 90.99 |
| Mean single sequence MFE | -38.92 |
| Consensus MFE | -30.72 |
| Energy contribution | -32.36 |
| Covariance contribution | 1.64 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.78 |
| Structure conservation index | 0.79 |
| SVM decision value | 2.26 |
| SVM RNA-class probability | 0.991218 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13518050 95 - 23771897 GCCGCCCCGAAAAGUGGCAUGCAACAUGCGAUUGCUGCCAAAAGUGCCGCCUGGCACGAGUCCAGGCUCAAGCGGCA-AGCACCAGGCGGCAGGCA (((...........(((((.(((((....).)))))))))....((((((((((...((((....))))..((....-.)).))))))))))))). ( -45.30) >DroSec_CAF1 5395 88 - 1 GCCGCCCCGAAAAGUGGCAUGCAACAUGCGAUUGCUGCCAAAAGUGCCGCCUGGCACGAGUCCAGGCCCAAGCGGCA-AGCACCAG-------GCA (((((.........(((((.(((((....).)))))))))........(((((((....).))))))....))))).-.((.....-------)). ( -32.90) >DroEre_CAF1 5367 95 - 1 GCCGCCCCGAAAAGUGGCAUGCAACAUGCGAUUGCUGCCAAAAGUGCCGCCUGGCACGAGUCCAGGCCCAAGCGGCA-AGCACCAGGCAGCAGGCA ((((((.(.....).))).(((.....)))..(((((((....((((.(((((((....).))))))((....))..-.))))..)))))))))). ( -40.40) >DroYak_CAF1 5375 96 - 1 GCCGCCCCGAAAAGUGGCAUGCAACAUGCGAUUGCUGCCAAAAGUGCCGCCUGGCACGAGUCCAGGCCCAAGCGGCAAAGCACCAGGCAGCAGGCA ((((((.(.....).))).(((.....)))..(((((((....((((.(((((((....).))))))((....))....))))..)))))))))). ( -40.40) >DroAna_CAF1 5656 87 - 1 GCCGCCCUGAAAAGUGGCAUGCAACAUGCGAUUGCUGCCAAAAGUGCCGCCUGGCACGCGUCCAGGACCAA--------GCACCAGGCAGCAAG-A (((((........))))).(((.....))).((((((((....((((..((((((....).))))).....--------))))..)))))))).-. ( -35.60) >consensus GCCGCCCCGAAAAGUGGCAUGCAACAUGCGAUUGCUGCCAAAAGUGCCGCCUGGCACGAGUCCAGGCCCAAGCGGCA_AGCACCAGGCAGCAGGCA (((((........))))).(((.....))).((((((((....((((.(((((((....).))))))((....))....))))..))))))))... (-30.72 = -32.36 + 1.64)


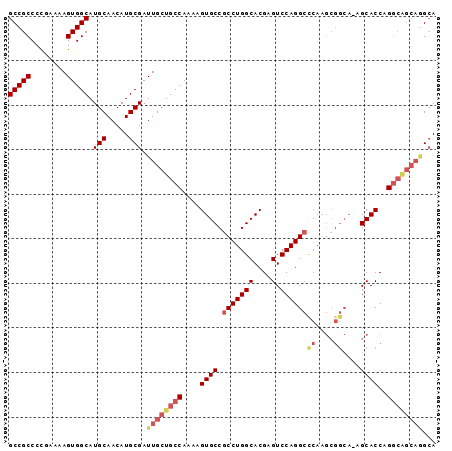
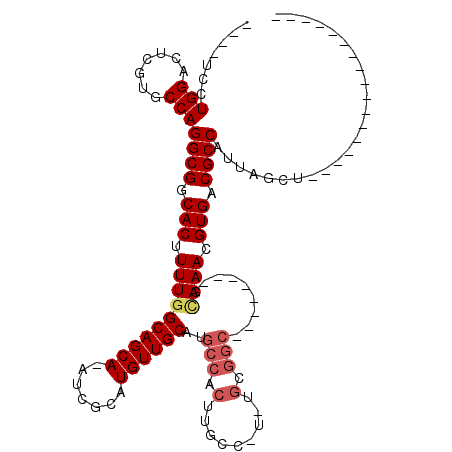
| Location | 13,518,074 – 13,518,168 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 74.62 |
| Mean single sequence MFE | -33.78 |
| Consensus MFE | -22.26 |
| Energy contribution | -22.98 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.830395 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13518074 94 + 23771897 UUGAGCCUGGACUCGUGCCAGGCGGCACUUUUGGCAGCA-AUCGCAUGUUGCAUGCCACUUUUC-GGGGCGGC--------ACAAACGUGACGCCAUUAGACGA--------------- ((((((((((.(....)))))))((((......((((((-......)))))).)))).....))-))((((.(--------((....))).)))).........--------------- ( -35.30) >DroVir_CAF1 17704 95 + 1 ----UUCUGGACUUGUGCCAGGCGGCACUUUUGGCAGCA-AUCGCAUGUUGCAUGCCACUUGCU-U-UGCGGCACGGUCCUGCAAACGUGACGCCAUUAGCU----------------- ----....(((((.(((((..((((((....((((((((-((.....))))).)))))..))))-.-.))))))))))))............((.....)).----------------- ( -32.00) >DroGri_CAF1 13000 99 + 1 GUGGUCCUGGAAUUGUGCCAGGCGGCACUUUUGGCAGCA-AUCGCAUGUUGCAUGCCACUUGCC-U-UGCGGCACGGUCCUGCAAACGUGACGCCAUUAGCU----------------- ((.(.(((((.......)))))).)).((..((((.((.-...))((((((((.(((...((((-.-...)))).)))..))).)))))...))))..))..----------------- ( -34.10) >DroWil_CAF1 7347 76 + 1 ----UCCUGGACUCGUGCCAGGCGGCACUUUUGGCAGCAAAUCGCAUGUUGCAUGCCACAC---------------------AAAACGUGACGCCAUUAG---A--------------- ----.(((((.(....)))))).(((.....(((((((((........)))).)))))(((---------------------.....)))..))).....---.--------------- ( -25.70) >DroMoj_CAF1 7338 94 + 1 ------UUGGACUUGUGCCAGGCGGCACUUUUGGCAGCA-AUCGCAUGUUGCAUGCCACUUGCUUU-UGCGGCAGGGUCCUGCAAACGUGACGCCAUUAGCU----------------- ------.(((.(....))))((((.(((...((((((((-((.....))))).)))))..(((...-.)))((((....))))....))).)))).......----------------- ( -33.20) >DroAna_CAF1 5672 109 + 1 UUGGUCCUGGACGCGUGCCAGGCGGCACUUUUGGCAGCA-AUCGCAUGUUGCAUGCCACUUUUC-AGGGCGGC--------GUAAACGUGACGCCAUUAGAUGGGCCUCGUCCUUUUUA ...((((((((...(((((....)))))...((((((((-((.....))))).)))))...)))-)))))(((--------((.......))))).......((((...))))...... ( -42.40) >consensus ____UCCUGGACUCGUGCCAGGCGGCACUUUUGGCAGCA_AUCGCAUGUUGCAUGCCACUUGCC_U_UGCGGC________GCAAACGUGACGCCAUUAGCU_________________ .......(((.......)))((((.(((.((((((((((.......))))))..(((.(.........).))).........)))).))).))))........................ (-22.26 = -22.98 + 0.72)


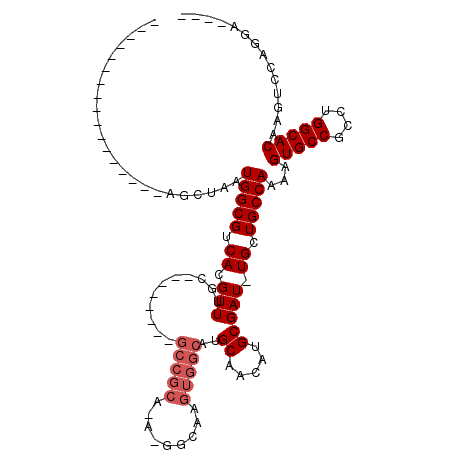
| Location | 13,518,074 – 13,518,168 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 74.62 |
| Mean single sequence MFE | -33.48 |
| Consensus MFE | -21.82 |
| Energy contribution | -22.65 |
| Covariance contribution | 0.83 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.65 |
| SVM decision value | 1.43 |
| SVM RNA-class probability | 0.953264 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13518074 94 - 23771897 ---------------UCGUCUAAUGGCGUCACGUUUGU--------GCCGCCCC-GAAAAGUGGCAUGCAACAUGCGAU-UGCUGCCAAAAGUGCCGCCUGGCACGAGUCCAGGCUCAA ---------------.........((((.(((....))--------).))))..-......(((((.(((((....).)-))))))))...(((((....)))))((((....)))).. ( -33.20) >DroVir_CAF1 17704 95 - 1 -----------------AGCUAAUGGCGUCACGUUUGCAGGACCGUGCCGCA-A-AGCAAGUGGCAUGCAACAUGCGAU-UGCUGCCAAAAGUGCCGCCUGGCACAAGUCCAGAA---- -----------------.((.((((......)))).)).((((.((((((..-.-.(((..(((((.(((((....).)-))))))))....)))....))))))..))))....---- ( -35.00) >DroGri_CAF1 13000 99 - 1 -----------------AGCUAAUGGCGUCACGUUUGCAGGACCGUGCCGCA-A-GGCAAGUGGCAUGCAACAUGCGAU-UGCUGCCAAAAGUGCCGCCUGGCACAAUUCCAGGACCAC -----------------.((.((((......)))).)).((.((.((((...-.-))))..(((((.(((((....).)-))))))))...(((((....))))).......)).)).. ( -37.20) >DroWil_CAF1 7347 76 - 1 ---------------U---CUAAUGGCGUCACGUUUU---------------------GUGUGGCAUGCAACAUGCGAUUUGCUGCCAAAAGUGCCGCCUGGCACGAGUCCAGGA---- ---------------.---.....((((.(((.((((---------------------(.(..((((((.....)))...)))..))))))))).))))((((....).)))...---- ( -24.80) >DroMoj_CAF1 7338 94 - 1 -----------------AGCUAAUGGCGUCACGUUUGCAGGACCCUGCCGCA-AAAGCAAGUGGCAUGCAACAUGCGAU-UGCUGCCAAAAGUGCCGCCUGGCACAAGUCCAA------ -----------------.((((..((((.(((....((((....)))).((.-...))...(((((.(((((....).)-))))))))...))).))))))))..........------ ( -32.40) >DroAna_CAF1 5672 109 - 1 UAAAAAGGACGAGGCCCAUCUAAUGGCGUCACGUUUAC--------GCCGCCCU-GAAAAGUGGCAUGCAACAUGCGAU-UGCUGCCAAAAGUGCCGCCUGGCACGCGUCCAGGACCAA ......(((((.(((.........(((((.......))--------)))(((((-....)).)))..(((((....).)-))).)))....(((((....))))).)))))........ ( -38.30) >consensus _________________AGCUAAUGGCGUCACGUUUGC________GCCGCA_A_GGCAAGUGGCAUGCAACAUGCGAU_UGCUGCCAAAAGUGCCGCCUGGCACAAGUCCAGGA____ .......................(((((.((.(((...........(((((.........)))))..((.....))))).)).)))))...(((((....))))).............. (-21.82 = -22.65 + 0.83)

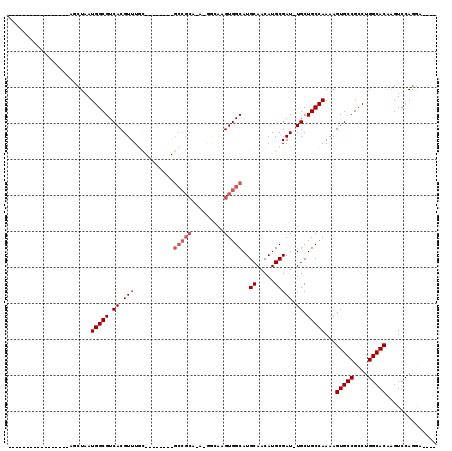
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:34:24 2006