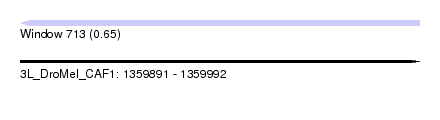
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 1,359,891 – 1,359,992 |
| Length | 101 |
| Max. P | 0.653203 |

| Location | 1,359,891 – 1,359,992 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.00 |
| Mean single sequence MFE | -30.33 |
| Consensus MFE | -20.48 |
| Energy contribution | -20.84 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.653203 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
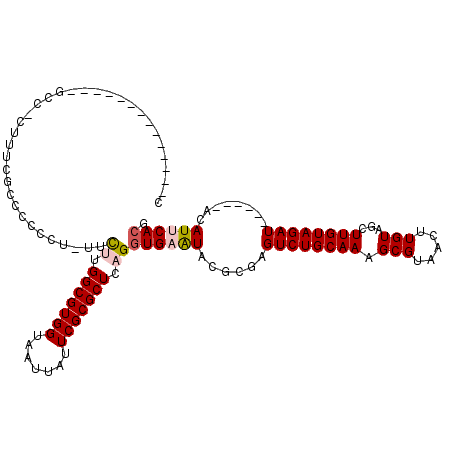
>3L_DroMel_CAF1 1359891 101 - 23771897 CU-------------UCCCCUUUUGCUCCCCUUUUCUUGGCGUGGUAAUUAUUCGCGCUCAGGUGAGUACGCGAGUCUGCAAAGCGUAACUUGUAGCUUGUAGAU------ACAUUCACG ..-------------........(((((.(((......(((((((.......))))))).))).))))).(((((.((((((........)))))))))))....------......... ( -26.40) >DroPse_CAF1 36312 113 - 1 CGUAUCUCGCAUCUCGCCUCUUUCCCCACGCU-CUUGUGGCGUGGUAAUUAUUCGCGCUCAGGUGAAUACGCGAGUCUGCAAAGCGUAACUUGUAACUUGUAGAU------ACAUUCACG .((((((((((.(((((........(((((((-.....)))))))....(((((((......))))))).)))))..)))...(((.....))).....).))))------))....... ( -35.30) >DroEre_CAF1 16153 105 - 1 C--------------GCCCCUUUCGCUCCCCU-UUCUUGGCGUGGUAAUUAUUCGCGCUCAGGUGAGUACGCGAGUCUGCAAAGCGUAACUUGUAGCUUGUAGAUACUCGUACAUCCACG .--------------.........((((.(((-.....(((((((.......))))))).))).))))..((((((((((((.((..........)))))))))..)))))......... ( -30.50) >DroYak_CAF1 16771 93 - 1 --------------------ACCUGCUCCCCU-UUCUUGGCGUGGUAAUUAUUCGCGCUCAGGUGAGUACGCGAGUCUGCAAAGCGUAACUUGUAGCUUGUAGAU------ACAUCCACG --------------------...(((((.(((-.....(((((((.......))))))).))).))))).(((((.((((((........)))))))))))....------......... ( -27.00) >DroAna_CAF1 15920 93 - 1 --------------------UCCUGCCUGCUU-UUCUUGGCGUGGUAAUUAUUCGCGCUUAGCUGAAUACGCGAGUCUGCAAAGCGUAACUUGUAGCUUGUAGAU------ACAUUCACG --------------------...((((((((.-.....)))).)))).....(((((............)))))((((((((.((..........))))))))))------......... ( -23.60) >DroPer_CAF1 37064 113 - 1 CGUAUCUCGCAUCUCGCCUCUUUCCCCACGCU-CUUGUGGCGUGGUAAGUAUUCGCGCUCAGGUGAAUACGCGAGUCUGCAAAGCGUAACUUGUAACUUGUAGAU------ACAUUCACG .((((((((((.(((((........(((((((-.....)))))))...((((((((......)))))))))))))..)))...(((.....))).....).))))------))....... ( -39.20) >consensus C______________GCC_CUUUCGCCCCCCU_UUCUUGGCGUGGUAAUUAUUCGCGCUCAGGUGAAUACGCGAGUCUGCAAAGCGUAACUUGUAGCUUGUAGAU______ACAUUCACG ...................................((.(((((((.......))))))).))((((((......((((((((.(((.....)))...))))))))........)))))). (-20.48 = -20.84 + 0.36)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:43:02 2006