
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,512,662 – 13,512,851 |
| Length | 189 |
| Max. P | 0.758303 |

| Location | 13,512,662 – 13,512,782 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.88 |
| Mean single sequence MFE | -32.13 |
| Consensus MFE | -30.52 |
| Energy contribution | -30.58 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.03 |
| Mean z-score | -0.83 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.574498 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
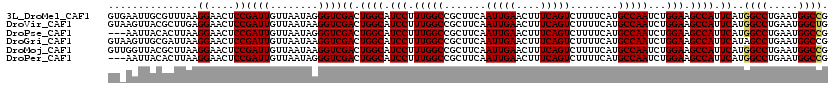
>3L_DroMel_CAF1 13512662 120 + 23771897 GUGAAUUGCGUUUAAGGAACUCCGAUUGUUAAUAGGGUCGACUGGCAUCCUUUGGCCGCUUCAAUUGAACUUUCAGUCUUUUCAUGCCAAUCUGGAAGCCAUUCAUGGCCUGAAUGGCCG ((((((...(((...((....))((((.(....).))))))).(((.(((.(((((.......(((((....)))))........)))))...))).)))))))))((((.....)))). ( -33.16) >DroVir_CAF1 10352 120 + 1 GUAAGUUACGCUUGAGGAACUCCGAUUGUUAAUAAGGUCGACUGGCAUCCUUUGGCCGCUUCAAUUGAACUUUCAGUCUUUUCAUGCCAAUCUGGAAGCCAUUCAUGGCCUGAAUGGCUG ..(((((.((.((((((.....(((((........)))))...(((........))).)))))).))))))).(((((..((((.((((...(((...)))....)))).)))).))))) ( -31.50) >DroPse_CAF1 2 117 + 1 ---AAUUACACUUAAGGAACUCCGAUUGUUAAUAGGGUCGACUGGCAUCCUUUGGCCGCUUCAAUUGAACUUUCAGUCUUUUCAUGCCAAUCUGGAAGCCAUUCAUGGCCUGAAUGGCCG ---............((....))((((.(....).))))((.((((.(((.(((((.......(((((....)))))........)))))...))).)))).))..((((.....)))). ( -32.56) >DroGri_CAF1 5436 120 + 1 GUAAGUUGCGAUUAAGGAACUCCGAUUGUUAAUAAGGUCGACUGGCAUCCUUUGGCCGCUUCAAUUGAACUUUCAGUCUUUUCAUGCCAAUCUGGAAGCCAUUCAUAGCCUGAAUGGCCG .......((.(((.(((......((((........))))((.((((.(((.(((((.......(((((....)))))........)))))...))).)))).))....))).))).)).. ( -29.96) >DroMoj_CAF1 15 120 + 1 GUUGGUUACGCUUAAGGAACUCCGAUUGUUAAUAAGGUCGACUGGCAUCCUUUGGCCGCUUCAAUUGAACUUUCAGUCUUUUCAUGCCAAUCUGGAAGCCAUUCAUGGCCUGAAUGGCCG ((..(((((.((((...(((.......)))..)))))).)))..)).(((.(((((.......(((((....)))))........)))))...))).((((((((.....)))))))).. ( -33.06) >DroPer_CAF1 2 117 + 1 ---AAUUACACUUAAGGAACUCCGAUUGUUAAUAGGGUCGACUGGCAUCCUUUGGCCGCUUCAAUUGAACUUUCAGUCUUUUCAUGCCAAUCUGGAAGCCAUUCAUGGCCUGAAUGGCCG ---............((....))((((.(....).))))((.((((.(((.(((((.......(((((....)))))........)))))...))).)))).))..((((.....)))). ( -32.56) >consensus GU_AAUUACGCUUAAGGAACUCCGAUUGUUAAUAAGGUCGACUGGCAUCCUUUGGCCGCUUCAAUUGAACUUUCAGUCUUUUCAUGCCAAUCUGGAAGCCAUUCAUGGCCUGAAUGGCCG ...............((....))((((........))))((.((((.(((.(((((.......(((((....)))))........)))))...))).)))).))..((((.....)))). (-30.52 = -30.58 + 0.06)
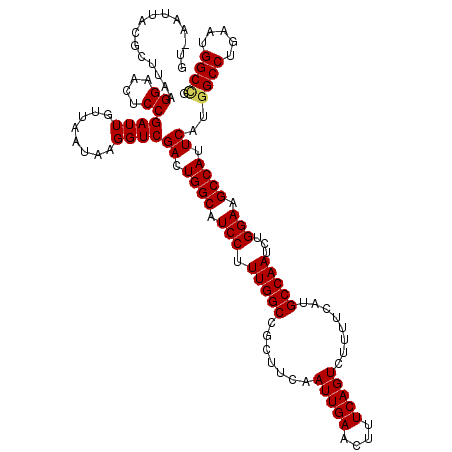

| Location | 13,512,662 – 13,512,782 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.88 |
| Mean single sequence MFE | -30.69 |
| Consensus MFE | -30.09 |
| Energy contribution | -29.95 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.03 |
| Mean z-score | -0.82 |
| Structure conservation index | 0.98 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.758303 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13512662 120 - 23771897 CGGCCAUUCAGGCCAUGAAUGGCUUCCAGAUUGGCAUGAAAAGACUGAAAGUUCAAUUGAAGCGGCCAAAGGAUGCCAGUCGACCCUAUUAACAAUCGGAGUUCCUUAAACGCAAUUCAC .((((.....)))).(((((.((.(((.(((((.........(((((....(((....)))(((.((...)).))))))))...........))))))))(((.....))))).))))). ( -30.65) >DroVir_CAF1 10352 120 - 1 CAGCCAUUCAGGCCAUGAAUGGCUUCCAGAUUGGCAUGAAAAGACUGAAAGUUCAAUUGAAGCGGCCAAAGGAUGCCAGUCGACCUUAUUAACAAUCGGAGUUCCUCAAGCGUAACUUAC .(((((((((.....)))))))))(((.(((((..((((...(((((....(((....)))(((.((...)).))))))))....))))...)))))))).................... ( -31.20) >DroPse_CAF1 2 117 - 1 CGGCCAUUCAGGCCAUGAAUGGCUUCCAGAUUGGCAUGAAAAGACUGAAAGUUCAAUUGAAGCGGCCAAAGGAUGCCAGUCGACCCUAUUAACAAUCGGAGUUCCUUAAGUGUAAUU--- .(((((((((.....)))))))))(((.(((((.........(((((....(((....)))(((.((...)).))))))))...........)))))))).................--- ( -29.95) >DroGri_CAF1 5436 120 - 1 CGGCCAUUCAGGCUAUGAAUGGCUUCCAGAUUGGCAUGAAAAGACUGAAAGUUCAAUUGAAGCGGCCAAAGGAUGCCAGUCGACCUUAUUAACAAUCGGAGUUCCUUAAUCGCAACUUAC .(((((((((.....)))))))))(((.(((((..((((...(((((....(((....)))(((.((...)).))))))))....))))...)))))))).................... ( -31.20) >DroMoj_CAF1 15 120 - 1 CGGCCAUUCAGGCCAUGAAUGGCUUCCAGAUUGGCAUGAAAAGACUGAAAGUUCAAUUGAAGCGGCCAAAGGAUGCCAGUCGACCUUAUUAACAAUCGGAGUUCCUUAAGCGUAACCAAC .(((((((((.....)))))))))(((.(((((..((((...(((((....(((....)))(((.((...)).))))))))....))))...)))))))).................... ( -31.20) >DroPer_CAF1 2 117 - 1 CGGCCAUUCAGGCCAUGAAUGGCUUCCAGAUUGGCAUGAAAAGACUGAAAGUUCAAUUGAAGCGGCCAAAGGAUGCCAGUCGACCCUAUUAACAAUCGGAGUUCCUUAAGUGUAAUU--- .(((((((((.....)))))))))(((.(((((.........(((((....(((....)))(((.((...)).))))))))...........)))))))).................--- ( -29.95) >consensus CGGCCAUUCAGGCCAUGAAUGGCUUCCAGAUUGGCAUGAAAAGACUGAAAGUUCAAUUGAAGCGGCCAAAGGAUGCCAGUCGACCCUAUUAACAAUCGGAGUUCCUUAAGCGUAACU_AC .(((((((((.....)))))))))(((.(((((.........(((((....(((....)))(((.((...)).))))))))...........)))))))).................... (-30.09 = -29.95 + -0.14)



| Location | 13,512,742 – 13,512,851 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 80.35 |
| Mean single sequence MFE | -28.45 |
| Consensus MFE | -19.81 |
| Energy contribution | -20.17 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.732313 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13512742 109 - 23771897 UUUGUCUAACGAGUUAUAUUUCA-U--UCU---UGGCAGGAUUUGUUUCAUUCGAUAAUCCCGCCAGCGCUCAAGCGGCCAUUCAGGCCAUGAAUGGCUUCCAGAUUGGCAUGAA ....((....)).......((((-(--..(---((((.(((((....((....)).))))).)))))(((....)))((((.((((((((....))))))...)).))))))))) ( -31.20) >DroVir_CAF1 10432 107 - 1 -AUUGCUAAC--AUUCAAUUUUGAA--AAU---UUGUAGGUUUUGUUUCAUUUGAUAACCCCACCAGCGCUCAAGCAGCCAUUCAGGCCAUGAAUGGCUUCCAGAUUGGCAUGAA -..((((...--.((((....))))--(((---(((..((..(((((......)))))..))....((......))(((((((((.....)))))))))..)))))))))).... ( -26.30) >DroGri_CAF1 5516 109 - 1 -UUGGCUAACGAAUCGAAUUAUGGU--AUU---UUAUAGGUUUUGUUUCAUUUGAUAACCCCACCAGCGCUCAAGCGGCCAUUCAGGCUAUGAAUGGCUUCCAGAUUGGCAUGAA -.................(((((..--...---.....((..(((((......)))))..)).(((((((....)))((((((((.....)))))))).......))))))))). ( -25.60) >DroMoj_CAF1 95 109 - 1 -AUUUCUAAU--AUUCAACUUUGUACAAUU---UUGUAGGUUUUGUUUCAUUUGAUAAUCCCACCAGCGCUCAAGCGGCCAUUCAGGCCAUGAAUGGCUUCCAGAUUGGCAUGAA -.........--.((((......((((...---.))))((..(((((......)))))..)).(((((((....)))((((((((.....)))))))).......))))..)))) ( -25.00) >DroAna_CAF1 95 108 - 1 -CUGUCAGAUUAAU---AUUGCC-C--UCUUUGUUGCAGGUUUUGUUUCAUUUGAUAAUCCCGCCAGCGCUCAAGCGGCCAUUCAAGCCAUGAAUGGCUUCCAGAUUGGCAUGAA -.((((((((.(((---(..(((-.--.(......)..)))..))))..)))))))).....((((((((....)))((((((((.....)))))))).......)))))..... ( -32.60) >DroPer_CAF1 79 107 - 1 UUUGCCUAAUCAA----AUGACU-C--UCUUU-AUGUAGGUUUCGUGUCCUUCGAUAAUCCAGCCAGCGCUCAAGCGGCCAUUCAGGCCAUGAAUGGCUUCCAGAUUGGCAUGAA ...(((((.....----((((..-.--...))-)).)))))((((((((..((..............(((....)))((((((((.....)))))))).....))..)))))))) ( -30.00) >consensus _UUGUCUAAC_AAUU_AAUUUCG_A__ACU___UUGUAGGUUUUGUUUCAUUUGAUAAUCCCACCAGCGCUCAAGCGGCCAUUCAGGCCAUGAAUGGCUUCCAGAUUGGCAUGAA ......................................((..(((((......)))))..)).(((((((....))(((((((((.....)))))))))....).))))...... (-19.81 = -20.17 + 0.36)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:34:16 2006