| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,496,437 – 13,496,528 |
| Length | 91 |
| Max. P | 0.698484 |

| Location | 13,496,437 – 13,496,528 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 76.01 |
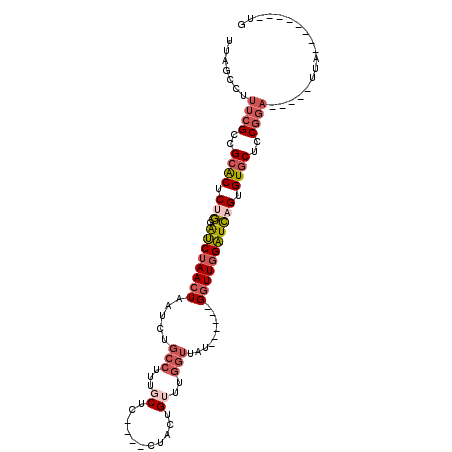
| Mean single sequence MFE | -25.10 |
| Consensus MFE | -15.30 |
| Energy contribution | -16.56 |
| Covariance contribution | 1.25 |
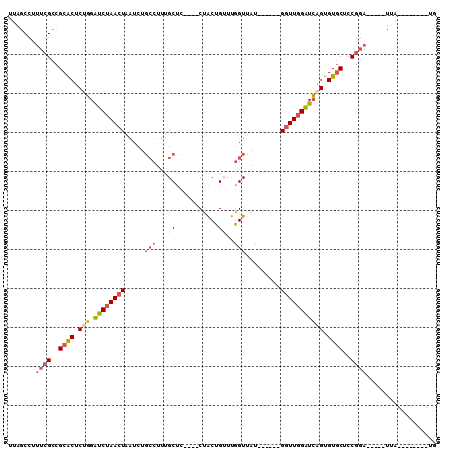
| Combinations/Pair | 1.17 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.698484 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
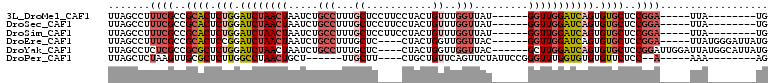
>3L_DroMel_CAF1 13496437 91 + 23771897 UUAGCCUUUCGCCGCACUCUGGAUCUAACUAAUCUGCCUUUGCUCCUUCCUACUGUUUGGUUAU------GGUUGGAUCAGUGUGCUCCGGA-----UUA--------UG .......((((..((((.(((.(((((((((....(((...((...........))..)))..)------))))))))))).))))..))))-----...--------.. ( -26.30) >DroSec_CAF1 8881 91 + 1 UUAGCCUUUCGCCGCACUCUGGAUCUAACUAAUCUGCCUUUGCUCCUUCCUACUGUUUGGUUAU------GGUUGGAUCAGUGUGCUCCGGA-----UUA--------UG .......((((..((((.(((.(((((((((....(((...((...........))..)))..)------))))))))))).))))..))))-----...--------.. ( -26.30) >DroSim_CAF1 9441 91 + 1 UUAGCCUUUCGCCGCACUCUGGAUCUAACUAAUCUGCCUUUGCUCCUUCCUACUGUUUGGUUAU------GGUUGGAUCAGUGUGCUCCGGA-----UUA--------UG .......((((..((((.(((.(((((((((....(((...((...........))..)))..)------))))))))))).))))..))))-----...--------.. ( -26.30) >DroEre_CAF1 8931 95 + 1 UUAGCCUUUCGCCGCACUCCGGAUCUAACUAAUCUGCCUUUGCUC----CUACUGGUUGGUUAC------GGUUGGAUCAGUGUGCUCCGGA-----UUAUGGGAUUAUG ....((.((((..((((....(((((((((((((.(((.......----.....))).))))..------)))))))))...))))..))))-----....))....... ( -27.20) >DroYak_CAF1 9270 100 + 1 UUAGCCUCUCGCCGCGCUCUGGAUCUAACUAAUCUGCCUUUGCUC----CUACUGGUUGGUUAC------GCUUGGAUCAGUGUGCUCCGGAUUGGAUUAUGGCAUUAUG ..........(((((((.(((.((((((((((((.(((.......----.....))).))))).------).))))))))).))))(((.....)))....)))...... ( -28.40) >DroPer_CAF1 10346 85 + 1 UUAGCUCUAAGUUGCGCUCUUGGCCUAACUGCU------UUGCUU----CUGCUGUUCAGUUCUAUUCCGGGUUUGGUGUGUGUUCUCC--A-----AAA--------AG .............((((.((..(((((((((..------..((..----..))....))))).......))))..)).)))).......--.-----...--------.. ( -16.11) >consensus UUAGCCUUUCGCCGCACUCUGGAUCUAACUAAUCUGCCUUUGCUC____CUACUGUUUGGUUAU______GGUUGGAUCAGUGUGCUCCGGA_____UUA________UG .......((((..((((.(((.((((((((.....(((...((...........))..))).........))))))))))).))))..)))).................. (-15.30 = -16.56 + 1.25)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:34:09 2006