
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,482,052 – 13,482,145 |
| Length | 93 |
| Max. P | 0.971380 |

| Location | 13,482,052 – 13,482,145 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 75.74 |
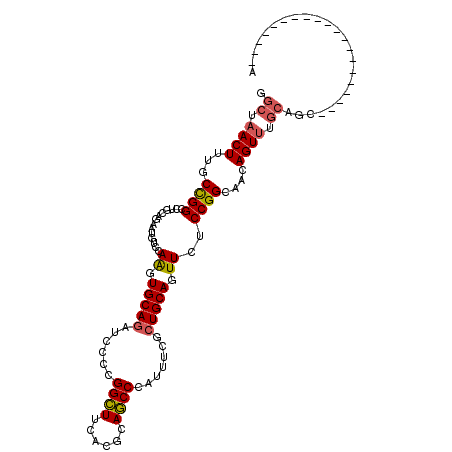
| Mean single sequence MFE | -31.26 |
| Consensus MFE | -17.23 |
| Energy contribution | -17.14 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.55 |
| SVM decision value | 1.67 |
| SVM RNA-class probability | 0.971380 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
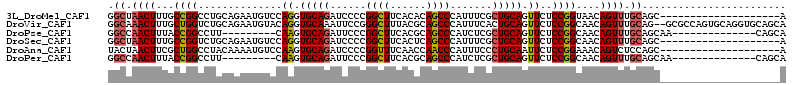
>3L_DroMel_CAF1 13482052 93 + 23771897 GGCUAACUUUGCCGGCCUGCAGAAUGUCCAGGUGCAGAUCCCCGGCUUCACACAGCCCAUUUCGCUGCAGUUCUCCGGUAACAGUUUGCAGC--------------------A .((.(((((((((((((((.........))))(((((......((((......)))).......))))).....))))))).)))).))...--------------------. ( -29.72) >DroVir_CAF1 21754 111 + 1 GGCAAACUUUGCUGGUCUGCAGAAUGUACAGGUGCAAAUUCCGGGCUUUACGCAGCCCAUUUCACUGCAGUUCUCCGGCAACAGUUUGCAG--GCGCCAGUGCAGGUGCAGCA .((((((((((((((.((((((..((((....))))......(((((......)))))......))))))....))))))).)))))))..--(((((......))))).... ( -49.80) >DroPse_CAF1 22865 90 + 1 GGCCAACUUUACCGGCCUU---------CAAGUGCAGAUUCCCGGCUUCACGCAGCCCAUCUCGCUGCAGUUCUCCGGCAACAGUUUGCAGCAA--------------CAGCA ((((.........))))..---------....((((((((.((((....(((((((.......))))).))...))))....))))))))((..--------------..)). ( -27.90) >DroSec_CAF1 15646 93 + 1 GGCUAACUUUGCCGGUCUGCAGAAUGUCCAGGUGCAGAUCCCCGGCUUCACUCAGCCCAUUUCGCUGCAGUUCUCCGGCAACAGUUUGCAGC--------------------A .((.(((((((((((.((((((..(((......)))((.....((((......))))....)).))))))....))))))).)))).))...--------------------. ( -33.00) >DroAna_CAF1 20845 93 + 1 UACUAACUUCGCUGGCCUACAAAAUGUCCAAGUGCAGAUCCCCGGUUUCAACCAACCCAUUUCCCUGCAAUUCUCCGGAAACAGUCUCCAGC--------------------A ..........(((((...((............(((((......((((......)))).......))))).......(....).))..)))))--------------------. ( -19.22) >DroPer_CAF1 26781 90 + 1 GGCCAACUUUACCGGCCUU---------CAAGUGCAGAUUCCCGGCUUCACGCAGCCCAUCUCGCUGCAGUUCUCCGGCAACAGUUUGCAGCAA--------------CAGCA ((((.........))))..---------....((((((((.((((....(((((((.......))))).))...))))....))))))))((..--------------..)). ( -27.90) >consensus GGCUAACUUUGCCGGCCUGCAGAAUGUCCAAGUGCAGAUCCCCGGCUUCACGCAGCCCAUUUCGCUGCAGUUCUCCGGCAACAGUUUGCAGC____________________A .((.((((...((((..............((.(((((......((((......)))).......))))).))..))))....)))).))........................ (-17.23 = -17.14 + -0.08)

| Location | 13,482,052 – 13,482,145 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 75.74 |
| Mean single sequence MFE | -32.58 |
| Consensus MFE | -23.57 |
| Energy contribution | -23.80 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.11 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.772985 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
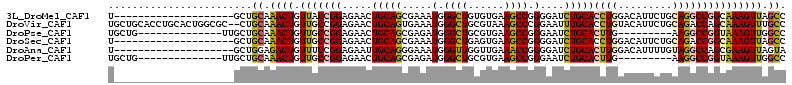
>3L_DroMel_CAF1 13482052 93 - 23771897 U--------------------GCUGCAAACUGUUACCGGAGAACUGCAGCGAAAUGGGCUGUGUGAAGCCGGGGAUCUGCACCUGGACAUUCUGCAGGCCGGCAAAGUUAGCC .--------------------((((((..(((....))).....)))))).....((.(((((.(((.(((((........)))))...)))))))).))(((.......))) ( -32.70) >DroVir_CAF1 21754 111 - 1 UGCUGCACCUGCACUGGCGC--CUGCAAACUGUUGCCGGAGAACUGCAGUGAAAUGGGCUGCGUAAAGCCCGGAAUUUGCACCUGUACAUUCUGCAGACCAGCAAAGUUUGCC ....((.((......)).))--..(((((((.((((.((....((((........(((((......)))))(((((.(((....))).))))))))).)).))))))))))). ( -39.50) >DroPse_CAF1 22865 90 - 1 UGCUG--------------UUGCUGCAAACUGUUGCCGGAGAACUGCAGCGAGAUGGGCUGCGUGAAGCCGGGAAUCUGCACUUG---------AAGGCCGGUAAAGUUGGCC .((..--------------..))((((....(((.((((...((.(((((.......)))))))....)))).))).))))....---------..((((((.....)))))) ( -29.50) >DroSec_CAF1 15646 93 - 1 U--------------------GCUGCAAACUGUUGCCGGAGAACUGCAGCGAAAUGGGCUGAGUGAAGCCGGGGAUCUGCACCUGGACAUUCUGCAGACCGGCAAAGUUAGCC .--------------------...((.((((.(((((((....(((((((.......)))(((((...(((((........))))).))))).)))).))))))))))).)). ( -36.90) >DroAna_CAF1 20845 93 - 1 U--------------------GCUGGAGACUGUUUCCGGAGAAUUGCAGGGAAAUGGGUUGGUUGAAACCGGGGAUCUGCACUUGGACAUUUUGUAGGCCAGCGAAGUUAGUA .--------------------(((((..((.((.((((.((...((((((....(.((((......)))).)...)))))))))))).))...))...))))).......... ( -27.40) >DroPer_CAF1 26781 90 - 1 UGCUG--------------UUGCUGCAAACUGUUGCCGGAGAACUGCAGCGAGAUGGGCUGCGUGAAGCCGGGAAUCUGCACUUG---------AAGGCCGGUAAAGUUGGCC .((..--------------..))((((....(((.((((...((.(((((.......)))))))....)))).))).))))....---------..((((((.....)))))) ( -29.50) >consensus U____________________GCUGCAAACUGUUGCCGGAGAACUGCAGCGAAAUGGGCUGCGUGAAGCCGGGAAUCUGCACCUGGACAUUCUGCAGGCCGGCAAAGUUAGCC ........................((.((((.(((((((.....(((((.....(.((((......)))).)....)))))((((.........))))))))))))))).)). (-23.57 = -23.80 + 0.23)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:34:05 2006