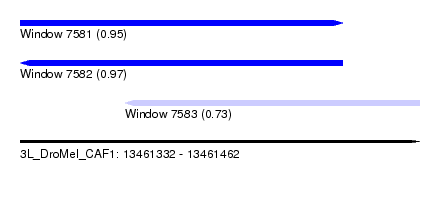
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,461,332 – 13,461,462 |
| Length | 130 |
| Max. P | 0.971466 |

| Location | 13,461,332 – 13,461,437 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 93.78 |
| Mean single sequence MFE | -27.82 |
| Consensus MFE | -25.25 |
| Energy contribution | -25.75 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.44 |
| SVM RNA-class probability | 0.953775 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13461332 105 + 23771897 UUGGUUUUGGUUUUGUUCCAUUUACAUAUAUGCCAAUGCCAGGCUCGGGCGAUAUAUCCAGUUUUUCACUUGGAUAUCUCGGUGUAUAUAUUGUAAGGCACACAU .............(((.((..(((((((((((((...(((.......)))((.((((((((........)))))))).))))))))))...))))))).)))... ( -26.90) >DroSec_CAF1 30356 104 + 1 UUGGUUUUGGUUUUGUUCCAUUUACAUAUAUGCCAAUGCCAGGCUCGGGCGAUAUAUCCAGUUUUUCACUUGGAUAUCUCGGUGUAUAUA-UGUAAGGCACACGU .............(((.((..(((((((((((((...(((.......)))((.((((((((........)))))))).)).).)))))))-))))))).)))... ( -29.10) >DroSim_CAF1 27734 104 + 1 UUGGUUUUGGUUUUGUUCCAUUUACAUAUAUGCCAAUGCCAGGCUCGGGCGAUAUAUCCAGUUUUUCACUUGGAUAUCUCGGUGUAUAUA-UGUAAGGCACACGU .............(((.((..(((((((((((((...(((.......)))((.((((((((........)))))))).)).).)))))))-))))))).)))... ( -29.10) >DroEre_CAF1 27888 96 + 1 -------UGGUUUUGUUCCAUUUACAUAUAUGCCAAUGCCAGGCUCGGGCGAUAUAUCCAGUUUUUCACUUGGAUAUCUCGGUGUAUAUA-UGUA-UGCACAUGC -------(((.......)))..((((((((((((...(((.......)))((.((((((((........)))))))).)).).)))))))-))))-......... ( -26.20) >consensus UUGGUUUUGGUUUUGUUCCAUUUACAUAUAUGCCAAUGCCAGGCUCGGGCGAUAUAUCCAGUUUUUCACUUGGAUAUCUCGGUGUAUAUA_UGUAAGGCACACGU .............(((.((..(((((((((((((...(((.......)))((.((((((((........)))))))).))))))))))...))))))).)))... (-25.25 = -25.75 + 0.50)

| Location | 13,461,332 – 13,461,437 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 93.78 |
| Mean single sequence MFE | -27.15 |
| Consensus MFE | -23.19 |
| Energy contribution | -23.25 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.46 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.68 |
| SVM RNA-class probability | 0.971466 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13461332 105 - 23771897 AUGUGUGCCUUACAAUAUAUACACCGAGAUAUCCAAGUGAAAAACUGGAUAUAUCGCCCGAGCCUGGCAUUGGCAUAUAUGUAAAUGGAACAAAACCAAAACCAA (((((((((................((.(((((((.((.....))))))))).))(((.......)))...))))))))).....(((.......)))....... ( -26.60) >DroSec_CAF1 30356 104 - 1 ACGUGUGCCUUACA-UAUAUACACCGAGAUAUCCAAGUGAAAAACUGGAUAUAUCGCCCGAGCCUGGCAUUGGCAUAUAUGUAAAUGGAACAAAACCAAAACCAA ...(((.(((((((-(((((....(((.(((((((.((.....))))))))).))).....(((.......)))))))))))))..)).)))............. ( -28.10) >DroSim_CAF1 27734 104 - 1 ACGUGUGCCUUACA-UAUAUACACCGAGAUAUCCAAGUGAAAAACUGGAUAUAUCGCCCGAGCCUGGCAUUGGCAUAUAUGUAAAUGGAACAAAACCAAAACCAA ...(((.(((((((-(((((....(((.(((((((.((.....))))))))).))).....(((.......)))))))))))))..)).)))............. ( -28.10) >DroEre_CAF1 27888 96 - 1 GCAUGUGCA-UACA-UAUAUACACCGAGAUAUCCAAGUGAAAAACUGGAUAUAUCGCCCGAGCCUGGCAUUGGCAUAUAUGUAAAUGGAACAAAACCA------- ((....)).-((((-(((((....(((.(((((((.((.....))))))))).))).....(((.......))))))))))))..(((.......)))------- ( -25.80) >consensus ACGUGUGCCUUACA_UAUAUACACCGAGAUAUCCAAGUGAAAAACUGGAUAUAUCGCCCGAGCCUGGCAUUGGCAUAUAUGUAAAUGGAACAAAACCAAAACCAA ..(((((((................((.(((((((.((.....))))))))).))(((.......)))...))))))).......(((.......)))....... (-23.19 = -23.25 + 0.06)



| Location | 13,461,366 – 13,461,462 |
|---|---|
| Length | 96 |
| Sequences | 3 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 95.10 |
| Mean single sequence MFE | -23.47 |
| Consensus MFE | -20.09 |
| Energy contribution | -20.43 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.726918 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13461366 96 - 23771897 UAAGCCGCACACACAUCUCUCUCGUAUGUGUGCCUUACAAUAUAUACACCGAGAUAUCCAAGUGAAAAACUGGAUAUAUCGCCCGAGCCUGGCAUU ...((((((((((.((.......)).)))))))................(((.(((((((.((.....))))))))).))).........)))... ( -26.20) >DroSec_CAF1 30390 94 - 1 CAAA-CGCACACACAUCUCUCGCGUACGUGUGCCUUACA-UAUAUACACCGAGAUAUCCAAGUGAAAAACUGGAUAUAUCGCCCGAGCCUGGCAUU ....-.((.((.....(((..(((((..((((.....))-))..)))...((.(((((((.((.....))))))))).))))..)))..))))... ( -22.10) >DroSim_CAF1 27768 94 - 1 CAAA-CGCACAAACAUCUCUCGCGUACGUGUGCCUUACA-UAUAUACACCGAGAUAUCCAAGUGAAAAACUGGAUAUAUCGCCCGAGCCUGGCAUU ....-.((.((.....(((..(((((..((((.....))-))..)))...((.(((((((.((.....))))))))).))))..)))..))))... ( -22.10) >consensus CAAA_CGCACACACAUCUCUCGCGUACGUGUGCCUUACA_UAUAUACACCGAGAUAUCCAAGUGAAAAACUGGAUAUAUCGCCCGAGCCUGGCAUU ......((((((...............)))))).................((.(((((((.((.....))))))))).))(((.......)))... (-20.09 = -20.43 + 0.33)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:34:01 2006