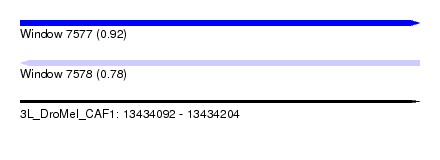
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,434,092 – 13,434,204 |
| Length | 112 |
| Max. P | 0.915681 |

| Location | 13,434,092 – 13,434,204 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 92.66 |
| Mean single sequence MFE | -25.80 |
| Consensus MFE | -23.16 |
| Energy contribution | -23.98 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.915681 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13434092 112 + 23771897 CAAGGACACUGUCACACAAAGGAGAACCCUCAUCAGUUGCAUGAAAUAAUUCCGUUUAAAUUAAGCGCCCCCCUGACAAA-AAGGAGCAGCAUUUCACGCGAUUCCCGUCCC--C ...((((..............(((....)))...((((((.((((((......(((((...)))))((..((((......-.))).)..)))))))).))))))...)))).--. ( -24.50) >DroSim_CAF1 33161 112 + 1 CAAGGACACUUUCACACAAAGGAGAACCCUCAUCAGUUGCAUGAAAUAAUUCCGUUUAAAUUAAGCGCCCUCCUGACAAA-AAGGAGCAGCAUUUCACGCGAUUCUCGUCCC--C ...((((.((((.....))))((((..........(((((.((((((......(((((...)))))((.(((((......-.)))))..)))))))).))))))))))))).--. ( -28.60) >DroEre_CAF1 32368 114 + 1 CAAGGACACUGACACACAAAGGAGUCCCCUCAUCCGUUGCAUGAAAUAAUUUCGUUUAAAUUAAGCUCCCUCCUGACAAA-AAGGAGCAGCAUUUUACGCGAUUCCCGUCCCCGC ...((((.............(((.........)))(((((.(((((((((((.....)))))).(((..(((((......-.))))).))).))))).)))))....)))).... ( -24.40) >DroYak_CAF1 31620 113 + 1 CAAGGACACUGUCACACAAAGGAGACCCCUCGUCAGUUGCAUGAAAUAAUUUCGUUUAAAUUAAGCGCCCUCCUCACAAAAAAGGAGCAGCAUUUCACGCCAUUCCCGUCCC--C ...((((((((.(.(.....)(((....)))).)))).((.((((((......(((((...)))))((.(((((........)))))..)))))))).)).......)))).--. ( -25.70) >consensus CAAGGACACUGUCACACAAAGGAGAACCCUCAUCAGUUGCAUGAAAUAAUUCCGUUUAAAUUAAGCGCCCUCCUGACAAA_AAGGAGCAGCAUUUCACGCGAUUCCCGUCCC__C ...((((..............(((....)))...((((((.((((((......(((((...)))))((.(((((........)))))..)))))))).))))))...)))).... (-23.16 = -23.98 + 0.81)

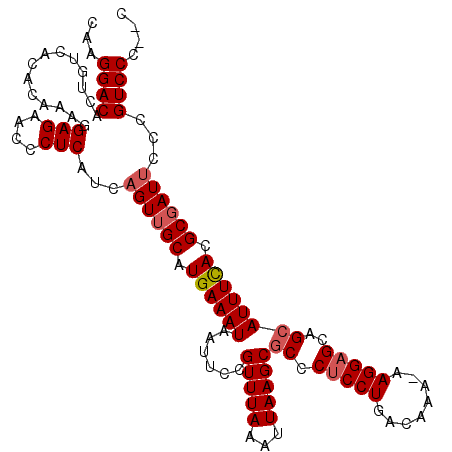
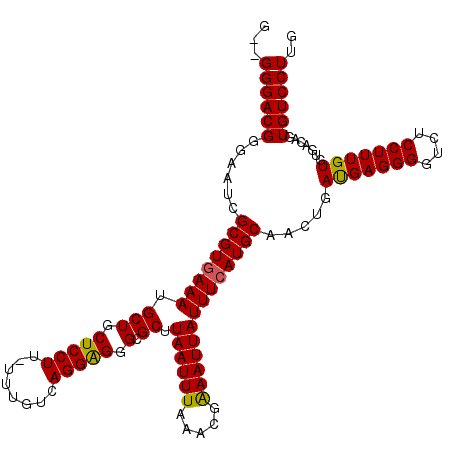
| Location | 13,434,092 – 13,434,204 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 92.66 |
| Mean single sequence MFE | -37.15 |
| Consensus MFE | -30.62 |
| Energy contribution | -30.25 |
| Covariance contribution | -0.38 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.783127 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13434092 112 - 23771897 G--GGGACGGGAAUCGCGUGAAAUGCUGCUCCUU-UUUGUCAGGGGGGCGCUUAAUUUAAACGGAAUUAUUUCAUGCAACUGAUGAGGGUUCUCCUUUGUGUGACAGUGUCCUUG (--(((((.......((((((((.((.(((((((-(.....)))))))))).((((((.....)))))))))))))).(((((..((((....))))..)....)))))))))). ( -37.30) >DroSim_CAF1 33161 112 - 1 G--GGGACGAGAAUCGCGUGAAAUGCUGCUCCUU-UUUGUCAGGAGGGCGCUUAAUUUAAACGGAAUUAUUUCAUGCAACUGAUGAGGGUUCUCCUUUGUGUGAAAGUGUCCUUG (--((((((((((((((((((((.(((.(((((.-......))))))))...((((((.....))))))))))))))..((....)))))))))((((.....)))).)))))). ( -35.70) >DroEre_CAF1 32368 114 - 1 GCGGGGACGGGAAUCGCGUAAAAUGCUGCUCCUU-UUUGUCAGGAGGGAGCUUAAUUUAAACGAAAUUAUUUCAUGCAACGGAUGAGGGGACUCCUUUGUGUGUCAGUGUCCUUG .((((((((.((...((((.((((...(((((((-((.....)))))))))...))))....(((.....))))))).(((((.(((....))).)))))...))..)))))))) ( -36.60) >DroYak_CAF1 31620 113 - 1 G--GGGACGGGAAUGGCGUGAAAUGCUGCUCCUUUUUUGUGAGGAGGGCGCUUAAUUUAAACGAAAUUAUUUCAUGCAACUGACGAGGGGUCUCCUUUGUGUGACAGUGUCCUUG (--(((((.......((((((((.(((.((((((......)))))))))...((((((.....)))))))))))))).(((((((((((....)))))))....)))))))))). ( -39.00) >consensus G__GGGACGGGAAUCGCGUGAAAUGCUGCUCCUU_UUUGUCAGGAGGGCGCUUAAUUUAAACGAAAUUAUUUCAUGCAACUGAUGAGGGGUCUCCUUUGUGUGACAGUGUCCUUG ...((((((......((((((((.(((.(((((........))))).).)).((((((.....)))))))))))))).....(((((((....))))))).......)))))).. (-30.62 = -30.25 + -0.38)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:33:56 2006