
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,433,685 – 13,433,882 |
| Length | 197 |
| Max. P | 0.988250 |

| Location | 13,433,685 – 13,433,802 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.00 |
| Mean single sequence MFE | -39.10 |
| Consensus MFE | -35.53 |
| Energy contribution | -36.52 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.62 |
| SVM RNA-class probability | 0.967751 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13433685 117 + 23771897 UCAUUACGGCAGCCAUCUUUUAUGGUGCCUCCGCCAAAGUAUCCAGCAUUAGUUUAUGUGUUAUUUGCCCACCCAUAUGGAUGUUGGCUUUUGGGGAGGCAGGGCGUAC---UACAUCAC ....((((.(.(((((.....)))))((((((.((((((...((((((((....((((.((.........)).))))..)))))))).))))))))))))..).)))).---........ ( -39.60) >DroSim_CAF1 32755 117 + 1 UCAUUACGGCAGCCAUCUUUUAUGGUGCCUCCGCCAAAGUAUCCAGCAUUAGUUUAUGUGUUAUUUGCCCACCCAUAUGGAUGUUGGCUUUUGGGGAGGCAGGGCGUAC---UACACACC ....((((.(.(((((.....)))))((((((.((((((...((((((((....((((.((.........)).))))..)))))))).))))))))))))..).)))).---........ ( -39.60) >DroEre_CAF1 31967 114 + 1 UCAUUACGGCAGCCAUCUUUUAUGGUGCCUCCGCCAAAGUAUCCAGCAUUAGUUUAUGUGUUAUUUGCCCACCCAUAUGGAUGUUGGCUUUUGGGGAGGCAGG------GCGUACAUACC ....((((.(.(((((.....)))))((((((.((((((...((((((((....((((.((.........)).))))..)))))))).))))))))))))..)------.))))...... ( -39.60) >DroYak_CAF1 31226 120 + 1 UCAUUACGGCAGCCAUCUUUUAUGGUGCCUCCGCCAAAGUAUCCAGCAUUAGUUUAUGUGUUAUUUGCCCACCCAUAUGGAUGUUGGCUUUUGGGGAGGCAGGACGUACGCGUACAUAGC ....((((...(((((.....)))))((((((.((((((...((((((((....((((.((.........)).))))..)))))))).))))))))))))....))))............ ( -37.60) >consensus UCAUUACGGCAGCCAUCUUUUAUGGUGCCUCCGCCAAAGUAUCCAGCAUUAGUUUAUGUGUUAUUUGCCCACCCAUAUGGAUGUUGGCUUUUGGGGAGGCAGGGCGUAC___UACAUACC ....((((.(.(((((.....)))))((((((.((((((...((((((((....((((.((.........)).))))..)))))))).))))))))))))..).))))............ (-35.53 = -36.52 + 1.00)



| Location | 13,433,685 – 13,433,802 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.00 |
| Mean single sequence MFE | -36.65 |
| Consensus MFE | -32.56 |
| Energy contribution | -33.12 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.51 |
| SVM RNA-class probability | 0.959891 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13433685 117 - 23771897 GUGAUGUA---GUACGCCCUGCCUCCCCAAAAGCCAACAUCCAUAUGGGUGGGCAAAUAACACAUAAACUAAUGCUGGAUACUUUGGCGGAGGCACCAUAAAAGAUGGCUGCCGUAAUGA .....(((---(.......((((((((((((.(((..(((((....))))))))......(((((......))).)).....))))).)))))))((((.....))))))))........ ( -35.10) >DroSim_CAF1 32755 117 - 1 GGUGUGUA---GUACGCCCUGCCUCCCCAAAAGCCAACAUCCAUAUGGGUGGGCAAAUAACACAUAAACUAAUGCUGGAUACUUUGGCGGAGGCACCAUAAAAGAUGGCUGCCGUAAUGA ((((((..---.)))))).((((((((((((.(((..(((((....))))))))......(((((......))).)).....))))).)))))))((((.....))))............ ( -37.50) >DroEre_CAF1 31967 114 - 1 GGUAUGUACGC------CCUGCCUCCCCAAAAGCCAACAUCCAUAUGGGUGGGCAAAUAACACAUAAACUAAUGCUGGAUACUUUGGCGGAGGCACCAUAAAAGAUGGCUGCCGUAAUGA ((((.......------..((((((((((((.(((..(((((....))))))))......(((((......))).)).....))))).)))))))((((.....)))).))))....... ( -36.60) >DroYak_CAF1 31226 120 - 1 GCUAUGUACGCGUACGUCCUGCCUCCCCAAAAGCCAACAUCCAUAUGGGUGGGCAAAUAACACAUAAACUAAUGCUGGAUACUUUGGCGGAGGCACCAUAAAAGAUGGCUGCCGUAAUGA ......((((.(((.....((((((((((((.(((..(((((....))))))))......(((((......))).)).....))))).)))))))((((.....)))).))))))).... ( -37.40) >consensus GGUAUGUA___GUACGCCCUGCCUCCCCAAAAGCCAACAUCCAUAUGGGUGGGCAAAUAACACAUAAACUAAUGCUGGAUACUUUGGCGGAGGCACCAUAAAAGAUGGCUGCCGUAAUGA ((((...............((((((((((((.(((..(((((....))))))))......(((((......))).)).....))))).)))))))((((.....)))).))))....... (-32.56 = -33.12 + 0.56)


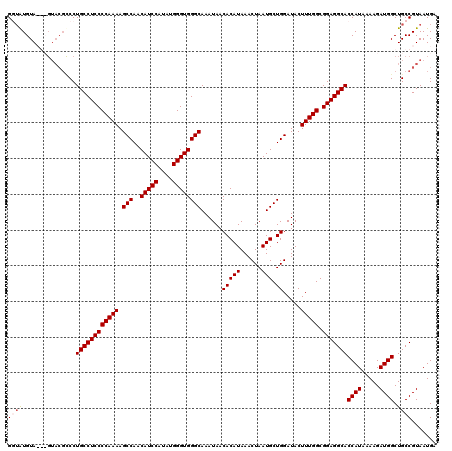
| Location | 13,433,725 – 13,433,842 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.58 |
| Mean single sequence MFE | -38.80 |
| Consensus MFE | -37.83 |
| Energy contribution | -37.45 |
| Covariance contribution | -0.38 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.97 |
| SVM decision value | 2.11 |
| SVM RNA-class probability | 0.988250 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13433725 117 - 23771897 AUGCAGGAGUGCCUCUGAUUUAUGCGGGGGCACAUACAUGGUGAUGUA---GUACGCCCUGCCUCCCCAAAAGCCAACAUCCAUAUGGGUGGGCAAAUAACACAUAAACUAAUGCUGGAU ..(((((.(((((((((.......))))))))).(((((....)))))---......)))))....(((...(((..(((((....))))))))........(((......))).))).. ( -38.40) >DroSim_CAF1 32795 117 - 1 AUGCAGGAGUGCCUCUGAUUUAUGCGGGGGCACAUACAUGGGUGUGUA---GUACGCCCUGCCUCCCCAAAAGCCAACAUCCAUAUGGGUGGGCAAAUAACACAUAAACUAAUGCUGGAU ..(((((((((((((((.......)))))))))...((.(((((((..---.)))))))))..)))......(((..(((((....))))))))..................)))..... ( -39.70) >DroEre_CAF1 32007 114 - 1 AUGCAGGAGUGCCUCUGAUUUAUGCGGGGGCACAUACGUGGGUAUGUACGC------CCUGCCUCCCCAAAAGCCAACAUCCAUAUGGGUGGGCAAAUAACACAUAAACUAAUGCUGGAU ..(((((.(((((((((.......)))))))((((((....))))))..))------)))))....(((...(((..(((((....))))))))........(((......))).))).. ( -38.00) >DroYak_CAF1 31266 120 - 1 AUGCAGGAGUGCCUCUGAUUUAUGCGGGGGCACAUACAUGGCUAUGUACGCGUACGUCCUGCCUCCCCAAAAGCCAACAUCCAUAUGGGUGGGCAAAUAACACAUAAACUAAUGCUGGAU ..(((((((((((((((.......))))))))).(((((....)))))........))))))....(((...(((..(((((....))))))))........(((......))).))).. ( -39.10) >consensus AUGCAGGAGUGCCUCUGAUUUAUGCGGGGGCACAUACAUGGGUAUGUA___GUACGCCCUGCCUCCCCAAAAGCCAACAUCCAUAUGGGUGGGCAAAUAACACAUAAACUAAUGCUGGAU ..(((((.(((((((((.......))))))))).(((((....))))).........)))))....(((...(((..(((((....))))))))........(((......))).))).. (-37.83 = -37.45 + -0.38)



| Location | 13,433,765 – 13,433,882 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.66 |
| Mean single sequence MFE | -40.45 |
| Consensus MFE | -38.03 |
| Energy contribution | -38.65 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.28 |
| SVM RNA-class probability | 0.939461 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13433765 117 - 23771897 CUCAAGACUUGUGAGAACAACCCAAGGACAAGGCUGGGUCAUGCAGGAGUGCCUCUGAUUUAUGCGGGGGCACAUACAUGGUGAUGUA---GUACGCCCUGCCUCCCCAAAAGCCAACAU .......((((.(........))))).....(((((((....(((((.(((((((((.......))))))))).(((((....)))))---......)))))...)))...))))..... ( -38.70) >DroSim_CAF1 32835 116 - 1 CUCAAGACUUGUGA-AACAACACAAGGACAAGGCUGGGUCAUGCAGGAGUGCCUCUGAUUUAUGCGGGGGCACAUACAUGGGUGUGUA---GUACGCCCUGCCUCCCCAAAAGCCAACAU .......((((((.-.....)))))).....(((((((....(((((.(((((((((.......))))))))).(((((....)))))---......)))))...)))...))))..... ( -43.60) >DroEre_CAF1 32047 114 - 1 CUUAAGACUUGUGAGAACAACACAAGGACGAGGCCGGGUCAUGCAGGAGUGCCUCUGAUUUAUGCGGGGGCACAUACGUGGGUAUGUACGC------CCUGCCUCCCCAAAAGCCAACAU .......((((((.......))))))...(((((.(((((((((.(..(((((((((.......)))))))))...)....)))))...))------)).)))))............... ( -41.60) >DroYak_CAF1 31306 120 - 1 CUUAAGACUUGCGAGAACAACACAAGGACGAGGCCGGGUCAUGCAGGAGUGCCUCUGAUUUAUGCGGGGGCACAUACAUGGCUAUGUACGCGUACGUCCUGCCUCCCCAAAAGCCAACAU .......((((.(.......).)))).....(((.(((....(((((((((((((((.......))))))))).(((((....)))))........))))))...)))....)))..... ( -37.90) >consensus CUCAAGACUUGUGAGAACAACACAAGGACAAGGCCGGGUCAUGCAGGAGUGCCUCUGAUUUAUGCGGGGGCACAUACAUGGGUAUGUA___GUACGCCCUGCCUCCCCAAAAGCCAACAU .......((((((.......)))))).....(((((((....(((((.(((((((((.......))))))))).(((((....))))).........)))))...)))...))))..... (-38.03 = -38.65 + 0.62)

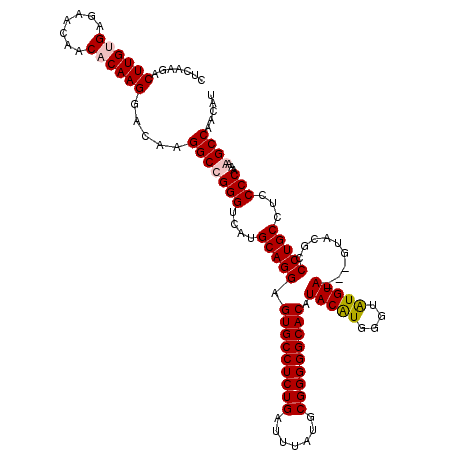

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:33:54 2006