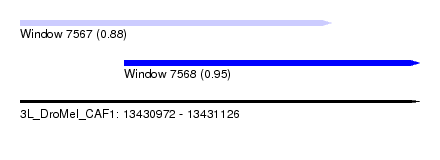
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,430,972 – 13,431,126 |
| Length | 154 |
| Max. P | 0.952500 |

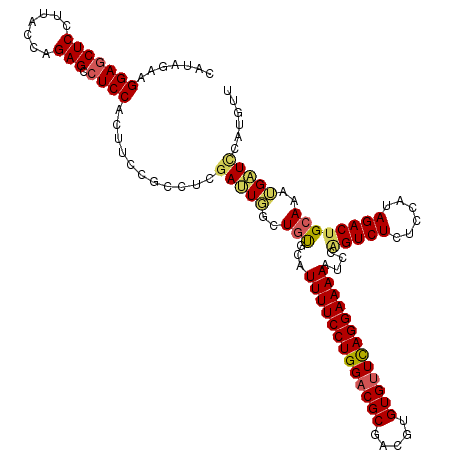
| Location | 13,430,972 – 13,431,092 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.25 |
| Mean single sequence MFE | -38.40 |
| Consensus MFE | -29.96 |
| Energy contribution | -30.24 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.877025 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13430972 120 + 23771897 CAUAGAAGGAGCUCCUUACCAGAGCCUCCACUUCCGCCUCGAUUGGCUGUGCAUUUUCCUGGACGCGACGUGUGUUCAGGAAAAAUCCAGUCUCUCCGUAGACUGAAAAUGUUUCAUGCA .......(((((((.......))).))))......(((......)))..(((((((((((((((((.....)))))))))))))...((((((......))))))...........)))) ( -41.30) >DroSec_CAF1 28139 120 + 1 CAUAGAAGGAGCUCCUUACCAGAGCCUCCACUUCCGCCUCGAUUGACUGUGCAUUUUCCUGGACGCGACGUGUGUUCAGGAAAAAUUCGGUCUCUCCAUAGACUGCAAAUGAUCCCAGUU .......(((((((.......))).))))...........((..(((((....(((((((((((((.....)))))))))))))...)))))..))....(((((..........))))) ( -35.60) >DroSim_CAF1 30007 120 + 1 CAUAGAAGGAACUCCUUACCAGAGCCUCCACUUCCGGCUCGAUUGGCUGUGCAUUUUCCUGGACGCGACGUGUGUUUAGGAAAAAUCCAGUCUCUCCAUAGACUGCAAAUGAUCCAAGUU (((((((((....)))).((((((((.........)))))...))))))))..(((((((((((((.....)))))))))))))...((((((......))))))............... ( -37.60) >DroEre_CAF1 29179 120 + 1 CAUAGAAGGAGCUCCUUACCAGAGCCUCCACCUCCGCCUCGACUUGCUGUGGAUUUUCCUGGACGCGGCCUGUGUUCAGGAAAAAUCCAGUCUCUCCAUAGACUGCAAAUGGUCCAUGUG .......(((((((.......))).)))).....(((...(((((((...((((((((((((((((.....))))))))).)))))))(((((......))))))))...))))...))) ( -41.30) >DroYak_CAF1 28386 120 + 1 CAUAGAAGGAGCUCCUUACCAGAGCCUCCACCUCGGCCUCAACUUGCUGCGGAUUUUCCUGGACGCGGCGUGUGCUCAGGAAAAAACCCGUCUCUCCAUAGACUGCAAAGGGUGCAUGUU .......(((((((.......))).)))).((.((((........)))).)).((((((((..(((.....)))..)))))))).((((((((......))))......))))....... ( -36.20) >consensus CAUAGAAGGAGCUCCUUACCAGAGCCUCCACUUCCGCCUCGAUUGGCUGUGCAUUUUCCUGGACGCGACGUGUGUUCAGGAAAAAUCCAGUCUCUCCAUAGACUGCAAAUGAUCCAUGUU .......(((((((.......))).))))...........(((((..(((...(((((((((((((.....)))))))))))))....(((((......))))))))..)))))...... (-29.96 = -30.24 + 0.28)

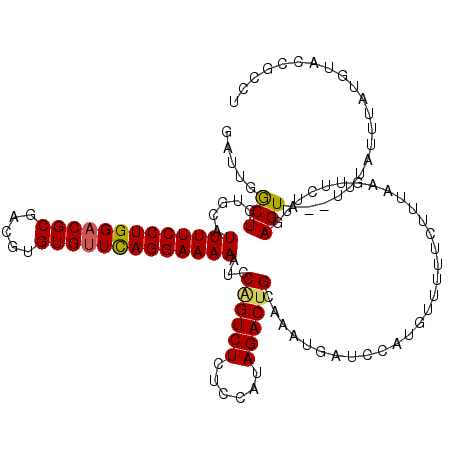
| Location | 13,431,012 – 13,431,126 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.71 |
| Mean single sequence MFE | -34.18 |
| Consensus MFE | -22.88 |
| Energy contribution | -22.84 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.67 |
| SVM decision value | 1.42 |
| SVM RNA-class probability | 0.952500 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13431012 114 + 23771897 GAUUGGCUGUGCAUUUUCCUGGACGCGACGUGUGUUCAGGAAAAAUCCAGUCUCUCCGUAGACUGAAAAUGUUUCAUGCA----UUUAAGUUCUUGAGUA--UUUAUUUAUGUACCGCCU ....(((.((((((((((((((((((.....)))))))))))))...((((((......)))))).((((((.....)))----))).............--........))))).))). ( -38.40) >DroSec_CAF1 28179 118 + 1 GAUUGACUGUGCAUUUUCCUGGACGCGACGUGUGUUCAGGAAAAAUUCGGUCUCUCCAUAGACUGCAAAUGAUCCCAGUUUAUCUUUAAGUUAUUGAGUA--UUUACUUAUGUACCGCCU (((.(((((....(((((((((((((.....)))))))))))))...((((((......))))))..........))))).))).....((...(((((.--...)))))...))..... ( -32.60) >DroSim_CAF1 30047 118 + 1 GAUUGGCUGUGCAUUUUCCUGGACGCGACGUGUGUUUAGGAAAAAUCCAGUCUCUCCAUAGACUGCAAAUGAUCCAAGUUUUUCUCUAAGUUCUUAAGUA--UUUAUUUAUGUAUCACCU ....((..((((((((((((((((((.....)))))))))))))...((((((......))))))...................................--........)))))..)). ( -27.40) >DroEre_CAF1 29219 118 + 1 GACUUGCUGUGGAUUUUCCUGGACGCGGCCUGUGUUCAGGAAAAAUCCAGUCUCUCCAUAGACUGCAAAUGGUCCAUGUGUUUCUUUAGAUUCUUGAGUAAAUCUAUUAAUG--GCGCCU (((((((...((((((((((((((((.....))))))))).)))))))(((((......))))))))...))))...(((((....((((((........)))))).....)--)))).. ( -38.20) >DroYak_CAF1 28426 116 + 1 AACUUGCUGCGGAUUUUCCUGGACGCGGCGUGUGCUCAGGAAAAAACCCGUCUCUCCAUAGACUGCAAAGGGUGCAUGUUUUUCUUUAGGUUCUUGAGUACAUUUAUUUAC----UGACU .....((((((..(......)..))))))((((((((((((....(((.((((......))))((((.....))))............)))))))))))))))........----..... ( -34.30) >consensus GAUUGGCUGUGCAUUUUCCUGGACGCGACGUGUGUUCAGGAAAAAUCCAGUCUCUCCAUAGACUGCAAAUGAUCCAUGUUUUUCUUUAAGUUCUUGAGUA__UUUAUUUAUGUACCGCCU .....(((.....(((((((((((((.....)))))))))))))...((((((......))))))...............................)))..................... (-22.88 = -22.84 + -0.04)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:33:47 2006