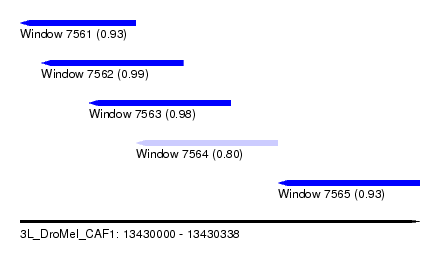
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,430,000 – 13,430,338 |
| Length | 338 |
| Max. P | 0.986285 |

| Location | 13,430,000 – 13,430,098 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.58 |
| Mean single sequence MFE | -33.56 |
| Consensus MFE | -28.62 |
| Energy contribution | -29.52 |
| Covariance contribution | 0.90 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.926691 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13430000 98 - 23771897 GAGCAAGUGCCAAAAUUGAUGUGCGUCAGGCGCUGGGCUCAUUCAUUAUUUUAAAAUCAAUUUAAAAGGUCUUGCCUUCCAAAGUUUUGACGCACGUC---------------------- (.((....)))......((((((((((((((..((((...........(((((((.....)))))))((.....)).))))..).)))))))))))))---------------------- ( -26.60) >DroSec_CAF1 27148 120 - 1 GAGCAAGUGCCAAAAUUGAUGUGCGUCAGGCGCUGGGCUCAUUCAUUAUUUUGAAAUCAAUUUAAAAGGUCUUGUCUUCCAAAGUUUUGACGCACGUCCUCCGCCUGGCAAGGGAAGCCA .......(((((.....((((((((((((((..((((..((.......(((((((.....))))))).....))...))))..).))))))))))))).......))))).((....)). ( -34.40) >DroSim_CAF1 29017 120 - 1 GAGCAAGUGCCAAAAUUGAUGUGCGUCAGGCGCUGGGCUCAUUCAUUAUUUUGAAAUCAAUUUAAAAGGUCUUGCCUUCCAAAGUUUUGACGCACGUCCUCCGCCUGGCAAGGGAAGCCA .......(((((.....((((((((((((((..(((.....((((......))))..........((((.....)))))))..).))))))))))))).......))))).((....)). ( -37.10) >DroEre_CAF1 28182 120 - 1 GAGCAAGUGCCAAAAUUGAUGUGCGUCAGGCGCUUGGCUCAUUCAUUAUUUUAAAAUCAAUUUAAAAGGUCUUGCCUUUCAAAGUUUUGACGCACGUCCUCCACUUGGCAAGGGAAGCCA .......((((((....(((((((((((((.(((((((.....(....(((((((.....))))))).)....))).....)))))))))))))))))......)))))).((....)). ( -35.30) >DroYak_CAF1 27396 120 - 1 GAGCAAGUGCCAAAAUUGAUGUGCGUCAGGCGCUGGGCUCAUUCAUUAUUUUAAAAUCAAUUUAAAAGGUCUUGCCUUUCAAAGUUUUGACGCACGUCCUUCACUUGGCAAGGAACGCCA .......((((((....(((((((((((((.(((((((.....(....(((((((.....))))))).)....)))).....))))))))))))))))......)))))).((....)). ( -34.40) >consensus GAGCAAGUGCCAAAAUUGAUGUGCGUCAGGCGCUGGGCUCAUUCAUUAUUUUAAAAUCAAUUUAAAAGGUCUUGCCUUCCAAAGUUUUGACGCACGUCCUCCACCUGGCAAGGGAAGCCA .......(((((.....(((((((((((((.(((((((.....(....(((((((.....))))))).)....)))).....)))))))))))))))).......))))).((....)). (-28.62 = -29.52 + 0.90)

| Location | 13,430,018 – 13,430,138 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.42 |
| Mean single sequence MFE | -34.72 |
| Consensus MFE | -33.72 |
| Energy contribution | -33.08 |
| Covariance contribution | -0.64 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.97 |
| SVM decision value | 2.04 |
| SVM RNA-class probability | 0.986285 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13430018 120 - 23771897 AUUGCUGCAAUGAUGGUUGAAAGAGGAGGCAUACGGCGAUGAGCAAGUGCCAAAAUUGAUGUGCGUCAGGCGCUGGGCUCAUUCAUUAUUUUAAAAUCAAUUUAAAAGGUCUUGCCUUCC .......((((....)))).....(((((((...((((((((((.((((((.....((((....))))))))))..))))))).....(((((((.....))))))).))).))))))). ( -35.40) >DroSec_CAF1 27188 120 - 1 AUUGCUGCAAUGAUGGUUGAAAGAGGAGGCAUACGGCGAUGAGCAAGUGCCAAAAUUGAUGUGCGUCAGGCGCUGGGCUCAUUCAUUAUUUUGAAAUCAAUUUAAAAGGUCUUGUCUUCC .......((((....)))).....(((((((...((((((((((.((((((.....((((....))))))))))..)))))))..(((..(((....)))..)))...))).))))))). ( -33.30) >DroSim_CAF1 29057 120 - 1 AUUGCUGCAAUGAUGGUUGAAAGAGGAGGCAUACGGCGAUGAGCAAGUGCCAAAAUUGAUGUGCGUCAGGCGCUGGGCUCAUUCAUUAUUUUGAAAUCAAUUUAAAAGGUCUUGCCUUCC .......((((....)))).....(((((((...((((((((((.((((((.....((((....))))))))))..)))))))..(((..(((....)))..)))...))).))))))). ( -36.00) >DroEre_CAF1 28222 120 - 1 AUUGCUGCAAUGAUGGUUCAAAGAGGAGGCAUAAGGCGAUGAGCAAGUGCCAAAAUUGAUGUGCGUCAGGCGCUUGGCUCAUUCAUUAUUUUAAAAUCAAUUUAAAAGGUCUUGCCUUUC ..........(((....)))....(((((((..((((((((((((((((((.....((((....))))))))))).))))))).....(((((((.....))))))).))))))))))). ( -34.90) >DroYak_CAF1 27436 120 - 1 AUUGCUGCAAUGAUGGUUGAAAGAGGAGGCAUAUGGCGAUGAGCAAGUGCCAAAAUUGAUGUGCGUCAGGCGCUGGGCUCAUUCAUUAUUUUAAAAUCAAUUUAAAAGGUCUUGCCUUUC .......((((....)))).....(((((((...((((((((((.((((((.....((((....))))))))))..))))))).....(((((((.....))))))).))).))))))). ( -34.00) >consensus AUUGCUGCAAUGAUGGUUGAAAGAGGAGGCAUACGGCGAUGAGCAAGUGCCAAAAUUGAUGUGCGUCAGGCGCUGGGCUCAUUCAUUAUUUUAAAAUCAAUUUAAAAGGUCUUGCCUUCC ........................(((((((...((((((((((.((((((.....((((....))))))))))..))))))).....(((((((.....))))))).))).))))))). (-33.72 = -33.08 + -0.64)

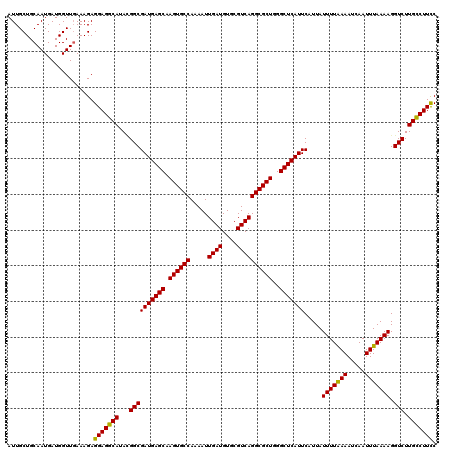
| Location | 13,430,058 – 13,430,178 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.42 |
| Mean single sequence MFE | -34.18 |
| Consensus MFE | -31.68 |
| Energy contribution | -31.88 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.93 |
| SVM RNA-class probability | 0.983141 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13430058 120 - 23771897 AACGAAUAAAGGAAACAAAGGAGCAACUUUGUCAUUAGUCAUUGCUGCAAUGAUGGUUGAAAGAGGAGGCAUACGGCGAUGAGCAAGUGCCAAAAUUGAUGUGCGUCAGGCGCUGGGCUC ..((......(....)......((..((((.(((...(((((((...)))))))...))).))))...))...)).....((((.((((((.....((((....))))))))))..)))) ( -34.00) >DroSec_CAF1 27228 120 - 1 AACGAAUAAAGGAAACAAAGGAGCAACUUUGUCAUUAGUCAUUGCUGCAAUGAUGGUUGAAAGAGGAGGCAUACGGCGAUGAGCAAGUGCCAAAAUUGAUGUGCGUCAGGCGCUGGGCUC ..((......(....)......((..((((.(((...(((((((...)))))))...))).))))...))...)).....((((.((((((.....((((....))))))))))..)))) ( -34.00) >DroSim_CAF1 29097 120 - 1 AACGAAUAAAGGAAACAAAGGAGCAACUUUGUCAUUAGUCAUUGCUGCAAUGAUGGUUGAAAGAGGAGGCAUACGGCGAUGAGCAAGUGCCAAAAUUGAUGUGCGUCAGGCGCUGGGCUC ..((......(....)......((..((((.(((...(((((((...)))))))...))).))))...))...)).....((((.((((((.....((((....))))))))))..)))) ( -34.00) >DroEre_CAF1 28262 120 - 1 AACGAAUGAAGGAAACAAAGGAGCAACUUUGUCAUUAGUCAUUGCUGCAAUGAUGGUUCAAAGAGGAGGCAUAAGGCGAUGAGCAAGUGCCAAAAUUGAUGUGCGUCAGGCGCUUGGCUC ..........(....)....((((..............((((((((......(((.(((......))).)))..)))))))).((((((((.....((((....)))))))))))))))) ( -34.90) >DroYak_CAF1 27476 120 - 1 AACGAAUAAAGGAAACAAAGGAGCAACUUUGUCAUUAGUCAUUGCUGCAAUGAUGGUUGAAAGAGGAGGCAUAUGGCGAUGAGCAAGUGCCAAAAUUGAUGUGCGUCAGGCGCUGGGCUC ..((.(((..(....)......((..((((.(((...(((((((...)))))))...))).))))...)).)))..))..((((.((((((.....((((....))))))))))..)))) ( -34.00) >consensus AACGAAUAAAGGAAACAAAGGAGCAACUUUGUCAUUAGUCAUUGCUGCAAUGAUGGUUGAAAGAGGAGGCAUACGGCGAUGAGCAAGUGCCAAAAUUGAUGUGCGUCAGGCGCUGGGCUC ..((......(....)......((..((((.(((...(((((((...)))))))...))).))))...))......))..((((.((((((.....((((....))))))))))..)))) (-31.68 = -31.88 + 0.20)

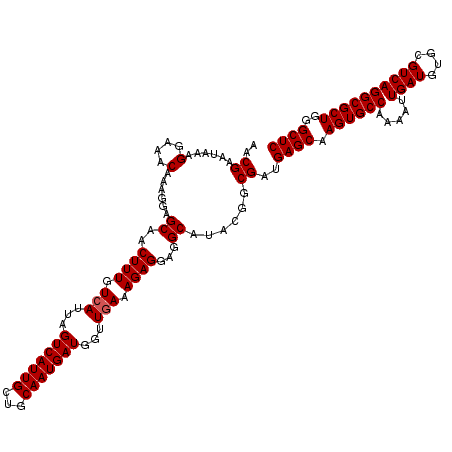
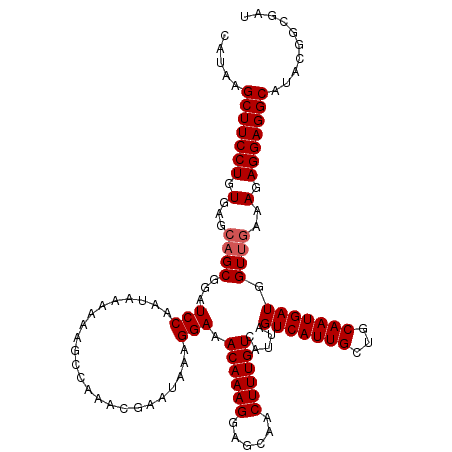
| Location | 13,430,098 – 13,430,218 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.42 |
| Mean single sequence MFE | -29.21 |
| Consensus MFE | -25.20 |
| Energy contribution | -25.60 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.801977 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13430098 120 - 23771897 CAUAAGCUUCCUGUAAGCUGCGGAUCCAAUAAAAAAGCCAAACGAAUAAAGGAAACAAAGGAGCAACUUUGUCAUUAGUCAUUGCUGCAAUGAUGGUUGAAAGAGGAGGCAUACGGCGAU .....(((((((.(..(((.(((..............))...........(....)...).))).......(((...(((((((...)))))))...))).).))))))).......... ( -26.94) >DroSec_CAF1 27268 120 - 1 CAUAAGCUUCCUGUGAGCAGCGGAUCCAAUAAAAAAGCCAAACGAAUAAAGGAAACAAAGGAGCAACUUUGUCAUUAGUCAUUGCUGCAAUGAUGGUUGAAAGAGGAGGCAUACGGCGAU .....(((((((.(...((((...(((.......................))).((((((......)))))).....(((((((...))))))).))))..).))))))).......... ( -27.60) >DroSim_CAF1 29137 119 - 1 CAUAAGCUUCCUUUGAGCAGCGGAUCCAAUAAA-AAGCCAAACGAAUAAAGGAAACAAAGGAGCAACUUUGUCAUUAGUCAUUGCUGCAAUGAUGGUUGAAAGAGGAGGCAUACGGCGAU .....(((((((((...((((...(((......-................))).((((((......)))))).....(((((((...))))))).))))..))))))))).......... ( -29.15) >DroEre_CAF1 28302 120 - 1 CAUAAGCUUCCUGUGAGCAGCGGAUCCAAUAAAAAAGCCAAACGAAUGAAGGAAACAAAGGAGCAACUUUGUCAUUAGUCAUUGCUGCAAUGAUGGUUCAAAGAGGAGGCAUAAGGCGAU .....(((((((.(..(((((((..............)).....(((((..((.((((((......))))))..))..))))))))))..(((....))).).))))))).......... ( -31.94) >DroYak_CAF1 27516 120 - 1 CAUAAGCUUCCUGUGAGCAGCGGAUCCAAUAAAAAAGCCAAACGAAUAAAGGAAACAAAGGAGCAACUUUGUCAUUAGUCAUUGCUGCAAUGAUGGUUGAAAGAGGAGGCAUAUGGCGAU ((((.(((((((.(...((((...(((.......................))).((((((......)))))).....(((((((...))))))).))))..).))))))).))))..... ( -30.40) >consensus CAUAAGCUUCCUGUGAGCAGCGGAUCCAAUAAAAAAGCCAAACGAAUAAAGGAAACAAAGGAGCAACUUUGUCAUUAGUCAUUGCUGCAAUGAUGGUUGAAAGAGGAGGCAUACGGCGAU .....(((((((.(...((((...(((.......................))).((((((......)))))).....(((((((...))))))).))))..).))))))).......... (-25.20 = -25.60 + 0.40)

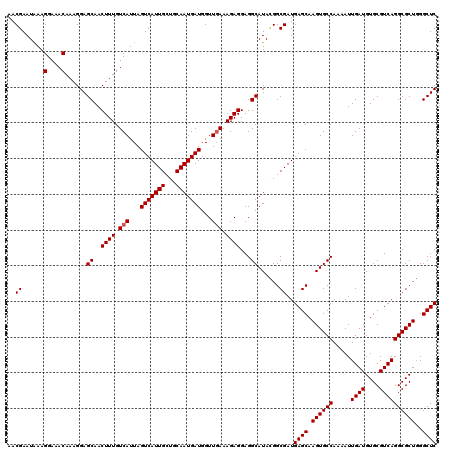
| Location | 13,430,218 – 13,430,338 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.65 |
| Mean single sequence MFE | -38.42 |
| Consensus MFE | -32.48 |
| Energy contribution | -32.08 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.932564 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13430218 120 - 23771897 CUCCAGCUGCCGCUGGUCCACACACAAAAUGGGCCAAAAACAGGAGGAGGAGCAGCCUGCCUCAGGCAUCUAUUUCCUGGAGACCCGACAGUCCUCACCCUUCUCCCGUGGAGCAUACUU .....(((.(((((((((((.........)))))))......((((((((....(((((...)))))..........((..(((......)))..)).)))))))).)))))))...... ( -43.80) >DroSec_CAF1 27388 117 - 1 CUCCAGCUGCCGCUGGUCCACCCACAAAAUGGGCCAAAAAUUGGA---GGAGCAGUCUGCCUCAGGCAUCUAUUUCCUGGAGACCCGACAGUCCUCACCCUUCUCCCGUGGAGCCUACUU .....(((.(((((((((((.........)))))))......(((---((((..(((((...)))))..........((..(((......)))..))..))))))).)))))))...... ( -36.00) >DroSim_CAF1 29256 117 - 1 CUCCAGCUGCCGCUGGUCCACCCACAAAAUGGGCCAAAAAUUGGA---GGAGCAGCCUGCCUCAGGCAUCUAUUUCCUGGAGACCCGACAGUCCUCACCCUUUUCCCGUGGAGCCUACUU .....(((.(((((((((((.........)))))))......(((---((((..(((((...)))))..........((..(((......)))..))..))))))).)))))))...... ( -36.20) >DroEre_CAF1 28422 117 - 1 CUCCAGCUGCCGCUGGUCCACCCACAAAAUGGGCCAAAAAGAGGA---GGAGCAGCCUGCCACUGGCAUCUAUUUCCUGGAGACUCGACAGUCCUCACCCUUCUCCCGUGGAGCCUACUU .....(((.(((((((((((.........)))))))....(((((---((....(((.......)))..........((..((((....))))..)).)))))))..)))))))...... ( -38.20) >DroYak_CAF1 27636 117 - 1 CUCCAGCUGCCGCUGGUCUACCCACAAAAUGGGCCAGAAAGAGGA---GGAGCAGCCUGCCUCGGGCAUCUAUUUCCUGGAGACUCGACAGUCCUCGCCCUACUCCCGUGGAGCCUACUU .....(((.((((.(((((.((((.....))))..)))..(((((---((.....))).))))((((.((((.....))))((((....))))...))))....)).)))))))...... ( -37.90) >consensus CUCCAGCUGCCGCUGGUCCACCCACAAAAUGGGCCAAAAAGAGGA___GGAGCAGCCUGCCUCAGGCAUCUAUUUCCUGGAGACCCGACAGUCCUCACCCUUCUCCCGUGGAGCCUACUU .....(((.(((((((((((.........)))))))......(((...(((...(((((...))))).)))...)))((..(((......)))..))..........)))))))...... (-32.48 = -32.08 + -0.40)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:33:43 2006