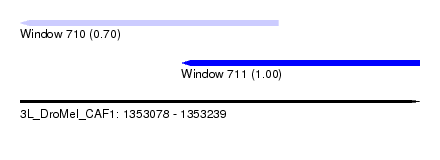
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 1,353,078 – 1,353,239 |
| Length | 161 |
| Max. P | 0.997092 |

| Location | 1,353,078 – 1,353,182 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.14 |
| Mean single sequence MFE | -25.04 |
| Consensus MFE | -12.29 |
| Energy contribution | -12.08 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.49 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.696643 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1353078 104 - 23771897 CAUCUCUUGGGACCAGUCAU-CUCUUUUGUAUCUGAACUUGUUAACAUAUCGAGUAUAAUUUUGAUGUGUUGUGGCCUCCU--------UCCAUUUC---CCAGCUCAAA--UAGCAA-- .......(((((.(((.((.-......))...))).......((((((((((((......)))))))))))).........--------......))---)))(((....--.)))..-- ( -22.00) >DroSec_CAF1 8591 107 - 1 CAUCUCUUGGGAGCAGUCAU-CUCUUUUGUAUCUCGACUUGUUAACAUAUCGAGUACGAUUUUGAUGUCUUGUGGCCU-----CCAUCCUCCAUCUC---CCUGCUCAAA--UAGCAA-- ........(((((.((.(((-(....(((((.(((((..((....))..))))))))))....)))).)).((((...-----)))).......)))---))((((....--.)))).-- ( -29.10) >DroSim_CAF1 6548 112 - 1 CAUCUCUUGGGAGCAGUCAU-CUCUUUUGUAUCUCGACUUGUUAACAUAUCGAGUACGAUUUUGAUGUCUUGUGGCCUCCU--CCAUCCUCCAUCUC---GCUGCUCAAA--UAGCAAAU ..........((((((((((-(....(((((.(((((..((....))..))))))))))....))))....((((......--))))..........---)))))))...--........ ( -28.90) >DroEre_CAF1 4468 108 - 1 CAUCUCCUGGGACUAGUCAU-CUCUUUUGUAUCUCGACUUGUUAACAUAUCGAGUACAAUUUUGAUGUGUUGUGGCCUUCU---------CCAUCUCCUCGCUGCUCAAA--CAGCAAAU ........((((.....(((-(....(((((.(((((..((....))..))))))))))....))))....((((......---------)))).)))).((((......--)))).... ( -26.00) >DroYak_CAF1 9609 105 - 1 CAUCUCUUGGGACUAGUCAU-CUCUUUUGUAUCUCGACUUGUUAACAUAUCGAGUACAAUUUUGAUGUGUGGUGGCCUUCU---------UCAUCUC---UCUGCUCAAA--UAGCAAAU ........((((.....(((-(....(((((.(((((..((....))..))))))))))....))))...(((((......---------)))))))---))((((....--.))))... ( -23.50) >DroAna_CAF1 8599 116 - 1 CAUCUCUUGGGACCCAUCUUGCUCUUUUGUCCCGCGACUUGUUAACAUAUUGAACACAUUUUUGAUAUGUUGUGGCCUCUCCUCGCUCC-UCGCCUU---UUUUCUCAAAACCGUCAAAU ........(((((...............)))))((((......(((((((..((......))..)))))))(..((........))..)-))))...---.................... ( -20.76) >consensus CAUCUCUUGGGACCAGUCAU_CUCUUUUGUAUCUCGACUUGUUAACAUAUCGAGUACAAUUUUGAUGUGUUGUGGCCUCCU_________CCAUCUC___CCUGCUCAAA__UAGCAAAU ......(((((((..((((.......(((((.(((((..((....))..))))))))))...))))..)).((((...............))))..........)))))........... (-12.29 = -12.08 + -0.22)

| Location | 1,353,143 – 1,353,239 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 81.26 |
| Mean single sequence MFE | -29.35 |
| Consensus MFE | -21.66 |
| Energy contribution | -24.94 |
| Covariance contribution | 3.28 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.10 |
| Structure conservation index | 0.74 |
| SVM decision value | 2.80 |
| SVM RNA-class probability | 0.997092 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1353143 96 - 23771897 --------CACGAGAUACGAGCUACCACCAGCCACGAGAUACUUAUUUAUGUGGGCUAUCUCUGGCAUCUCUUGGGACCAGUCAUCUCUUUUGUAUCUGAACUU --------....(((((((((.........((((.(((((((((((....))))).)))))))))).......((((.......)))).)))))))))...... ( -26.90) >DroSec_CAF1 8659 93 - 1 --------CACGAGAUACCAGCUAG---CAGCCACGAGAUACUUAUUUAUGUGGGCUAUCUCUGGCAUCUCUUGGGAGCAGUCAUCUCUUUUGUAUCUCGACUU --------..((((((((.......---..((((.(((((((((((....))))).)))))))))).......((((.......))))....)))))))).... ( -31.00) >DroSim_CAF1 6621 72 - 1 --------CAC------------------------GAGAUACUUAUUUAUGUGGGCUAUCUCUGGCAUCUCUUGGGAGCAGUCAUCUCUUUUGUAUCUCGACUU --------..(------------------------(((((((...........((((.((((.((.....)).))))..)))).........)))))))).... ( -18.95) >DroEre_CAF1 4537 101 - 1 AUGCGAGAUACGAGAUGCGAGCUAC---CAGCCACGAGAUACUUAUUUAUGUGGGCUAUCUCUGGCAUCUCCUGGGACUAGUCAUCUCUUUUGUAUCUCGACUU ...((((((((((((((...((((.---..((((.(((((((((((....))))).)))))))))).(((....))).))))))))))....)))))))).... ( -37.20) >DroYak_CAF1 9675 101 - 1 CGCCCAGAUACGAGAUACGAGCUAC---CAGCCACGAGAUACUUAUUUAUGUGGGCUAUCUCUGGCAUCUCUUGGGACUAGUCAUCUCUUUUGUAUCUCGACUU ..........(((((((((((....---..((((.(((((((((((....))))).)))))))))).......((((.......)))).))))))))))).... ( -32.70) >consensus ________CACGAGAUACGAGCUAC___CAGCCACGAGAUACUUAUUUAUGUGGGCUAUCUCUGGCAUCUCUUGGGACCAGUCAUCUCUUUUGUAUCUCGACUU ..........(((((((((((.........((((.(((((((((((....))))).)))))))))).......((((.......)))).))))))))))).... (-21.66 = -24.94 + 3.28)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:43:00 2006