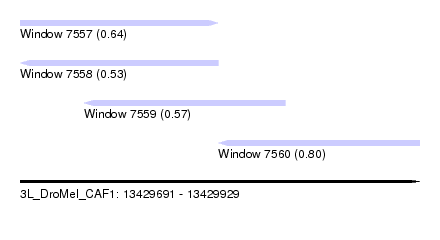
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,429,691 – 13,429,929 |
| Length | 238 |
| Max. P | 0.800461 |

| Location | 13,429,691 – 13,429,809 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.58 |
| Mean single sequence MFE | -19.32 |
| Consensus MFE | -16.68 |
| Energy contribution | -15.76 |
| Covariance contribution | -0.92 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.640158 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13429691 118 + 23771897 --UUUCCAUGGUAUUCCCCUUGUUUCAAUAAAAACCAUUUAAUAAUACGAUUUAUCUGAAGUUAUCACGAAGCUCACGCACUCCCCAUUGAGUUCUCCUUCAGUGGCAUAAAUCUGCCUC --.......((((........((((......)))).............(((((((((((((..........((....))((((......))))....))))))....))))))))))).. ( -17.30) >DroSec_CAF1 26835 120 + 1 UAUUUCCAUGGUAUUCCCCUUGUUUUAAUAAAAACCAUUUAAUAAUACGAUUUAUCUGAAUUUAUCACGAAGCUCACGCACUCCCCAUUGAGUUCUCCUUCAGUGGCAUAAAUCUGCCUC .........((((........((.(((.((((.....)))).))).))(((((((................((....)).....((((((((......)))))))).))))))))))).. ( -18.00) >DroSim_CAF1 28704 120 + 1 UAUUUCCAUGGUAUUCCCCUUGGUUUAAUAAAAACCAUUUAAUAAUACGAUUUAUCUGAAGUUAUCACGAAGCUCACGCACUCCCCAUUGAGUUCUCCUUCAGUGGCAUAAAUCUGCCUC .......((.(((((.....((((((.....))))))......))))).))....((((((..........((....))((((......))))....)))))).((((......)))).. ( -22.60) >DroEre_CAF1 27869 120 + 1 UAUUACCAUGGUAUUCCCCUUGAUUUAACGAAAUCCAUUUAAUAACAAGAUUUAUCUGAAGUUAUCACGAAGCUCACGCACUCCCCAUUGAGUUCUCCUUCAAUGGCAUAAAUCUGCCUC .........(((.........(((((....)))))............((((((((................((....)).....((((((((......)))))))).))))))))))).. ( -18.70) >DroYak_CAF1 27076 120 + 1 GAUUACCAUGGUAUUCCCCUGGCUUUAACCAAAUCCAUUAAAUAAUAAGAUUUAUCUGAAGUUAUCACGAAGCUCACGCACUCCCCAUUGAGUUCUCCUUCAAUGGCAUAAAUCUGCCUC ((.(((....))).))....((((((((.........))))).....((((((((................((....)).....((((((((......)))))))).))))))))))).. ( -20.00) >consensus UAUUUCCAUGGUAUUCCCCUUGUUUUAAUAAAAACCAUUUAAUAAUACGAUUUAUCUGAAGUUAUCACGAAGCUCACGCACUCCCCAUUGAGUUCUCCUUCAGUGGCAUAAAUCUGCCUC .........((((......(((......))).................(((((((................((....)).....((((((((......)))))))).))))))))))).. (-16.68 = -15.76 + -0.92)



| Location | 13,429,691 – 13,429,809 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.58 |
| Mean single sequence MFE | -28.80 |
| Consensus MFE | -22.78 |
| Energy contribution | -23.22 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.79 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.525976 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13429691 118 - 23771897 GAGGCAGAUUUAUGCCACUGAAGGAGAACUCAAUGGGGAGUGCGUGAGCUUCGUGAUAACUUCAGAUAAAUCGUAUUAUUAAAUGGUUUUUAUUGAAACAAGGGGAAUACCAUGGAAA-- ((((((......)))).((((((....((((......))))((....))..........)))))).....))......((..(((((.(((.(((...)))...))).)))))..)).-- ( -27.90) >DroSec_CAF1 26835 120 - 1 GAGGCAGAUUUAUGCCACUGAAGGAGAACUCAAUGGGGAGUGCGUGAGCUUCGUGAUAAAUUCAGAUAAAUCGUAUUAUUAAAUGGUUUUUAUUAAAACAAGGGGAAUACCAUGGAAAUA ..((((......)))).(((((.....((((......))))((....))...........)))))..(((((((........))))))).........((.((......)).))...... ( -24.20) >DroSim_CAF1 28704 120 - 1 GAGGCAGAUUUAUGCCACUGAAGGAGAACUCAAUGGGGAGUGCGUGAGCUUCGUGAUAACUUCAGAUAAAUCGUAUUAUUAAAUGGUUUUUAUUAAACCAAGGGGAAUACCAUGGAAAUA ..((((......)))).((((((....((((......))))((....))..........))))))..(((((((........)))))))........(((.((......)).)))..... ( -30.60) >DroEre_CAF1 27869 120 - 1 GAGGCAGAUUUAUGCCAUUGAAGGAGAACUCAAUGGGGAGUGCGUGAGCUUCGUGAUAACUUCAGAUAAAUCUUGUUAUUAAAUGGAUUUCGUUAAAUCAAGGGGAAUACCAUGGUAAUA ..(((((((((((.(((((((.(.....))))))))(((((((....)).........)))))..))))))).))))((((.(((((((((.((......)).))))).))))..)))). ( -31.00) >DroYak_CAF1 27076 120 - 1 GAGGCAGAUUUAUGCCAUUGAAGGAGAACUCAAUGGGGAGUGCGUGAGCUUCGUGAUAACUUCAGAUAAAUCUUAUUAUUUAAUGGAUUUGGUUAAAGCCAGGGGAAUACCAUGGUAAUC ..((((......)))).((((((....((((......))))((....))..........)))))).(((((((...........)))))))......((((.((.....)).)))).... ( -30.30) >consensus GAGGCAGAUUUAUGCCACUGAAGGAGAACUCAAUGGGGAGUGCGUGAGCUUCGUGAUAACUUCAGAUAAAUCGUAUUAUUAAAUGGUUUUUAUUAAAACAAGGGGAAUACCAUGGAAAUA ..((((......)))).((((((....((((......))))((....))..........)))))).................(((((.(((.((.......)).))).)))))....... (-22.78 = -23.22 + 0.44)


| Location | 13,429,729 – 13,429,849 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.00 |
| Mean single sequence MFE | -31.18 |
| Consensus MFE | -29.42 |
| Energy contribution | -29.58 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.19 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.572281 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13429729 120 - 23771897 CUUUGAGUGUGCUUCAAGGAUUUGCUUAUAUAUCCGAAUUGAGGCAGAUUUAUGCCACUGAAGGAGAACUCAAUGGGGAGUGCGUGAGCUUCGUGAUAACUUCAGAUAAAUCGUAUUAUU ...(((((.((((((((((((..........))))...)))))))).))))).....((((((....((((......))))((....))..........))))))............... ( -31.80) >DroSec_CAF1 26875 120 - 1 CUUUGAGUGUGCUUCAAGGAUUUGCUUAUAUAUCCGAAUUGAGGCAGAUUUAUGCCACUGAAGGAGAACUCAAUGGGGAGUGCGUGAGCUUCGUGAUAAAUUCAGAUAAAUCGUAUUAUU ...(((((.((((((((((((..........))))...)))))))).))))).....(((((.....((((......))))((....))...........)))))............... ( -28.50) >DroSim_CAF1 28744 120 - 1 CUUUGAGUGUGCUUCAAGGAUUUGCUUAUAUAUCCGAAUUGAGGCAGAUUUAUGCCACUGAAGGAGAACUCAAUGGGGAGUGCGUGAGCUUCGUGAUAACUUCAGAUAAAUCGUAUUAUU ...(((((.((((((((((((..........))))...)))))))).))))).....((((((....((((......))))((....))..........))))))............... ( -31.80) >DroEre_CAF1 27909 120 - 1 CUUUGAGUGUGCUUCAAGGAUUUGCUUAUAUAUCCGAAUUGAGGCAGAUUUAUGCCAUUGAAGGAGAACUCAAUGGGGAGUGCGUGAGCUUCGUGAUAACUUCAGAUAAAUCUUGUUAUU ...(((((.((((((((((((..........))))...)))))))).)))))..(((((((.(.....)))))))).(((.((....)))))..((((((...(((....))).)))))) ( -33.50) >DroYak_CAF1 27116 120 - 1 CUUUGAGUGUGCUUCAAGGAUUUGCUUAUAUAUCCGAAUCGAGGCAGAUUUAUGCCAUUGAAGGAGAACUCAAUGGGGAGUGCGUGAGCUUCGUGAUAACUUCAGAUAAAUCUUAUUAUU ....((((.(.((((((((((..........)))).......((((......)))).)))))).)..))))(((((((((.((....))))).(((.....))).......))))))... ( -30.30) >consensus CUUUGAGUGUGCUUCAAGGAUUUGCUUAUAUAUCCGAAUUGAGGCAGAUUUAUGCCACUGAAGGAGAACUCAAUGGGGAGUGCGUGAGCUUCGUGAUAACUUCAGAUAAAUCGUAUUAUU ...(((((.(((((((.((((..........))))....))))))).))))).....((((((....((((......))))((....))..........))))))............... (-29.42 = -29.58 + 0.16)


| Location | 13,429,809 – 13,429,929 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.50 |
| Mean single sequence MFE | -32.24 |
| Consensus MFE | -30.60 |
| Energy contribution | -30.68 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.800461 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13429809 120 - 23771897 CCUUUCCAUAUGCCAGAUGUUGUCUGAAGUGACGAUCCAGUCAACGUAUGGAAAAGGAAAACUUUAUCGGCAAUCGGCUCCUUUGAGUGUGCUUCAAGGAUUUGCUUAUAUAUCCGAAUU ((((((((((((.(((((...)))))...((((......)))).))))))).))))).........((((..((.((((((((.(((....))).)))))...))).))....))))... ( -34.20) >DroSec_CAF1 26955 120 - 1 CCUUUCCAUAUGCCAGAUGUUGUCUGAAGUGACGAUCCAGUCAACGUAUGGAAAAGGAAAAGUUUAUCGGCAAUCGGCUCCUUUGAGUGUGCUUCAAGGAUUUGCUUAUAUAUCCGAAUU ((((((((((((.(((((...)))))...((((......)))).))))))).))))).........((((..((.((((((((.(((....))).)))))...))).))....))))... ( -34.20) >DroSim_CAF1 28824 120 - 1 CCUUUCCAUAUGCCAGAUGUUGUCUGAAGUGACGAUCCAGUCAACGUAUGGAAAAGGAAAAGUUUAUCGGCAAUCGGCUCCUUUGAGUGUGCUUCAAGGAUUUGCUUAUAUAUCCGAAUU ((((((((((((.(((((...)))))...((((......)))).))))))).))))).........((((..((.((((((((.(((....))).)))))...))).))....))))... ( -34.20) >DroEre_CAF1 27989 120 - 1 CCUUUCCAUAUGCCAGAUGUUGUCUGAAGUGACGAUCCAGUCAACGCACGGAAGAAGAAAAGUUUAUCGGCAAUCGGCUCCUUUGAGUGUGCUUCAAGGAUUUGCUUAUAUAUCCGAAUU ..(((((...((((((((...)))))...((((......))))..))).)))))............((((..((.((((((((.(((....))).)))))...))).))....))))... ( -26.60) >DroYak_CAF1 27196 120 - 1 CCUUUCCAUAUGCCAGAUGUUGUCUGAAGUGACGAUCCAGUCAACGUAUGGAGAAAGAAAAGUUUAUCGGCAAUCGGCUCCUUUGAGUGUGCUUCAAGGAUUUGCUUAUAUAUCCGAAUC ..((((((((((.(((((...)))))...((((......)))).))))))))))............((((..((.((((((((.(((....))).)))))...))).))....))))... ( -32.00) >consensus CCUUUCCAUAUGCCAGAUGUUGUCUGAAGUGACGAUCCAGUCAACGUAUGGAAAAGGAAAAGUUUAUCGGCAAUCGGCUCCUUUGAGUGUGCUUCAAGGAUUUGCUUAUAUAUCCGAAUU ..((((((((((.(((((...)))))...((((......)))).))))))))))............((((..((.((((((((.(((....))).)))))...))).))....))))... (-30.60 = -30.68 + 0.08)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:33:39 2006