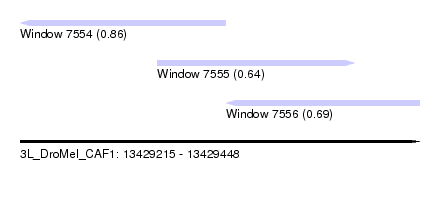
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,429,215 – 13,429,448 |
| Length | 233 |
| Max. P | 0.864932 |

| Location | 13,429,215 – 13,429,335 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.33 |
| Mean single sequence MFE | -31.46 |
| Consensus MFE | -26.10 |
| Energy contribution | -27.10 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.864932 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13429215 120 - 23771897 CGAAAUGCACUCGAUAGCAGUCCAAAAAGCACCCACCGCCCAGAAAAAUCGCACCCUGGUGUGUGCGUGUCACUUGAAAGUCGCGCUAAAGGAAAACUGUCACAAAUUGCAUUUAAAUGC ..(((((((...(((((...(((.....((((.(((((....((....))......))))).))))((((.(((....))).))))....)))...)))))......)))))))...... ( -32.10) >DroSec_CAF1 26338 120 - 1 CGAAAUGCACUGGAUGGCAGUCCAAAAAGCACCCACCACCCAGAAAAAUCGCACCCUGGUGUGUGCGUGUCACUUGAAAGUCGCGCUAAAGGAAAACUGUCACAAAUUGCAUUUAAAUGC ..(((((((...(.(((((((((.....((((.(((((....((....))......))))).))))((((.(((....))).))))....)))...)))))))....)))))))...... ( -34.80) >DroSim_CAF1 28206 120 - 1 CGAAAUGCACUGGAUAGCAGUCCAAAAAGCACCCACCACCCAGAAAAAUCGCACCCUGGUGUGUGCGUGUCACUUGAAAGUCGCGCUAAAGGAAAACUGUCACAAAUUGCAUUUAAAUGC ..(((((((.(((((((...(((.....((((.(((((....((....))......))))).))))((((.(((....))).))))....)))...))))).))...)))))))...... ( -34.90) >DroEre_CAF1 27362 119 - 1 CCAAAUGCACUGGAAAGCAUUU-GGGCUGCACCCACCACCCAGAAAAAUCGCACCCAGCUGUGUGCGUGUCACUUGAAAGUCGCGCUAAAGGAAAACUGUCACAAAUUGCAUUUAAAUGC ..(((((((((((...((...(-(((.((.......))))))((....))))..)))).((((.(((((.(........).)))))...((.....))..))))...)))))))...... ( -26.80) >DroYak_CAF1 26590 120 - 1 CCAAAUGCAGUGGAUAGCAGCCCAAAAAGCACCCGCCACCCAGAAAAAUCGCACCCUGCUGUGUGCGUGUCAGUUGAAAGUCGCGCUAAAGGAAAACUGUCACAAAUUGCAUUUAAAUGC ..((((((((((.(((((((........((....))......((....)).....))))))).)))(((.(((((.....((.(......))).))))).)))....)))))))...... ( -28.70) >consensus CGAAAUGCACUGGAUAGCAGUCCAAAAAGCACCCACCACCCAGAAAAAUCGCACCCUGGUGUGUGCGUGUCACUUGAAAGUCGCGCUAAAGGAAAACUGUCACAAAUUGCAUUUAAAUGC ..(((((((.(((((((...(((.....((((.(((((....((....))......))))).))))((((.(((....))).))))....)))...))))).))...)))))))...... (-26.10 = -27.10 + 1.00)


| Location | 13,429,295 – 13,429,410 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.25 |
| Mean single sequence MFE | -39.56 |
| Consensus MFE | -30.14 |
| Energy contribution | -30.30 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.638743 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13429295 115 + 23771897 GGCGGUGGGUGCUUUUUGGACUGCUAUCGAGUGCAUUUCGAGGAGUAAUAUGGGGGUGGC-----CACCCGGUUUGGAAAAAAGUUAAGGCAUGCCGUCGCACAAAGGAAUUUUCCUAUA .(((..((((((((((..(((((((.(((((.....)))))..))).......((((...-----.))))))))..))).........))))).))..)))....((((....))))... ( -36.20) >DroSec_CAF1 26418 115 + 1 GGUGGUGGGUGCUUUUUGGACUGCCAUCCAGUGCAUUUCGAGGAGCCAUAUGGGGGUGGC-----CACCCGGUUUGGAAAGAAGUUAAGGCAUGCCAUCGCACAAAGGAAUUUUCCUAUA .((((((((((((((...((((....(((((......(((.((.(((((......)))))-----..))))).)))))....))))))))))).)))))))....((((....))))... ( -37.60) >DroSim_CAF1 28286 115 + 1 GGUGGUGGGUGCUUUUUGGACUGCUAUCCAGUGCAUUUCGAGGAGCAAUAUGGGGGUGGC-----CACCCGGUUUGGAAAAAAGUUAAGGCAUGCCGUCGCACAAAGGAAUUUUCCUAUA .(((..((.(((((((((((.(((........))).))))))))))).((((.((((...-----.))))..(((....)))........))))))..)))....((((....))))... ( -34.50) >DroEre_CAF1 27442 118 + 1 GGUGGUGGGUGCAGCCC-AAAUGCUUUCCAGUGCAUUUGGAGGAGCAAUGUGGGGUUGGGGCCACCACCCAGUAUGGAUA-AAGUUAAGGCAUGCCGUCGCACAAAGGAAUUUUCCUAUA ((((((((.(.((((((-.(.((((((((((.....))))).))))).)...)))))).).))))))))..((((((...-..((....))...)))).))....((((....))))... ( -45.80) >DroYak_CAF1 26670 114 + 1 GGUGGCGGGUGCUUUUUGGGCUGCUAUCCACUGCAUUUGGAGGAGCAAUGUGGGGAUGGG-----CGCCCACUAUGGAAA-AAGUUAAGGCAUGCCGUCGCACAAAGGAAUUUUCCUAUA .((((((((((((((..((((..((((((.(..((((.(......)))))..))))))).-----.))))(((.(....)-.))).))))))).)))))))....((((....))))... ( -43.70) >consensus GGUGGUGGGUGCUUUUUGGACUGCUAUCCAGUGCAUUUCGAGGAGCAAUAUGGGGGUGGC_____CACCCGGUUUGGAAA_AAGUUAAGGCAUGCCGUCGCACAAAGGAAUUUUCCUAUA .(((((((.(((((((((((.(((........))).)))))))))))......(((((.......)))))........................)))))))....((((....))))... (-30.14 = -30.30 + 0.16)


| Location | 13,429,335 – 13,429,448 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.71 |
| Mean single sequence MFE | -32.83 |
| Consensus MFE | -24.30 |
| Energy contribution | -25.42 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.685548 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13429335 113 - 23771897 CCAAAAUGGUAGUUGUUGCGCAUAU--UUCCGUGCAAUAUUAUAGGAAAAUUCCUUUGUGCGACGGCAUGCCUUAACUUUUUUCCAAACCGGGUG-----GCCACCCCCAUAUUACUCCU .......(((((((((((((((..(--((((.............))))).......))))))))))).))))..................(((.(-----(...)))))........... ( -28.02) >DroSec_CAF1 26458 113 - 1 CCGGAAUGGUAGUUGUUGCGCAUAU--UUCCGUGCAAUAUUAUAGGAAAAUUCCUUUGUGCGAUGGCAUGCCUUAACUUCUUUCCAAACCGGGUG-----GCCACCCCCAUAUGGCUCCU ((((..(((.((..((((.((((.(--(..((..(((......((((....)))))))..))..)).))))..))))..))..)))..))))(..-----((((........))))..). ( -32.10) >DroSim_CAF1 28326 113 - 1 CCGGAAUGGUAGUUGUUGCGCAUAU--UUCCGUGCAAUAUUAUAGGAAAAUUCCUUUGUGCGACGGCAUGCCUUAACUUUUUUCCAAACCGGGUG-----GCCACCCCCAUAUUGCUCCU ..(((..(((((((((((((((..(--((((.............))))).......))))))))))).))))..................(((.(-----(...))))).......))). ( -31.02) >DroEre_CAF1 27481 119 - 1 CCGGCAUGGUAGUUGUUGGGCAUAUUUUUCCGUGCAAUGUUAUAGGAAAAUUCCUUUGUGCGACGGCAUGCCUUAACUU-UAUCCAUACUGGGUGGUGGCCCCAACCCCACAUUGCUCCU ((((.((((.(((((..((((((......(((((((.......((((....))))...))).)))).))))))))))).-...)))).))))((((.((......))))))......... ( -37.80) >DroYak_CAF1 26710 112 - 1 CUGGAAUGGAAGUUGUUGCGCAUAU--UUCCGUGCAAUAUUAUAGGAAAAUUCCUUUGUGCGACGGCAUGCCUUAACUU-UUUCCAUAGUGGGCG-----CCCAUCCCCACAUUGCUCCU ..((((((((((..((((.((((..--..(((((((.......((((....))))...))).)))).))))..))))..-))))))).(((((..-----......))))).....))). ( -35.20) >consensus CCGGAAUGGUAGUUGUUGCGCAUAU__UUCCGUGCAAUAUUAUAGGAAAAUUCCUUUGUGCGACGGCAUGCCUUAACUU_UUUCCAAACCGGGUG_____GCCACCCCCAUAUUGCUCCU ..(((..(((((((((((((((.....((((.............))))........))))))))))).))))..................(((((.......))))).........))). (-24.30 = -25.42 + 1.12)

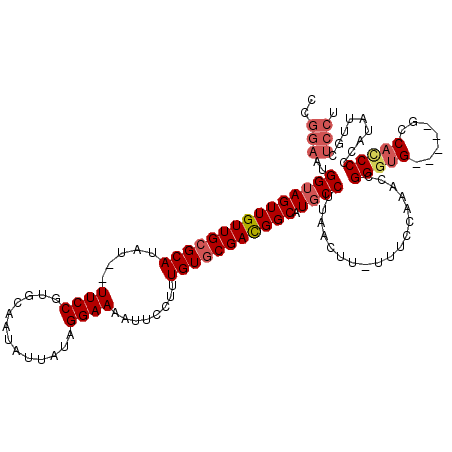
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:33:35 2006