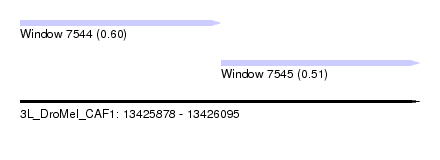
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,425,878 – 13,426,095 |
| Length | 217 |
| Max. P | 0.597987 |

| Location | 13,425,878 – 13,425,987 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.93 |
| Mean single sequence MFE | -22.74 |
| Consensus MFE | -17.94 |
| Energy contribution | -18.14 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.597987 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13425878 109 + 23771897 GCCAACAGUGCGUAUGACAGUUCUUACGCAAUUAAAAUACCAUCA---CUGGCUGUAAGAUUUACAUAAAUUCGGCCUGCGA-AACUGA-------AAACUGAUAAGCGUCAUAAAAAAA ((...(((((((((.((....)).))))).............((.---(.(((((...(((((....)))))))))).).))-......-------..))))....))............ ( -20.20) >DroSec_CAF1 22892 110 + 1 GCCAACAGUGCGUAUGACAGUUCUUACGCAAUUAAAAUACCAUCAUCGCUGGCUGUAAGAUUUACAUAAAUUCGGCCUGCGA-AACUGA-------AAACUGAUAAGCGUCAUAAAA--A ((...(((((((((.((....)).))))).............(((((((.(((((...(((((....)))))))))).))))-...)))-------..))))....)).........--. ( -25.90) >DroSim_CAF1 24666 117 + 1 GCCAACAGUGCGUAUGACAGUUCUUACGCAAUUAAAAUACCAUCAUCGCUGGCUGUAAGAUUUACAUAAAUUCGGCCUGCGA-AACUGAAAACUGAAAACUGAUAAGCGUCAUAAAA--A ((...(((((((((.((....)).))))).............(((((((.(((((...(((((....)))))))))).))))-...))).........))))....)).........--. ( -25.90) >DroEre_CAF1 24077 106 + 1 GCC-ACAGUGCGUAUGACAGUUCUUACGCAAUUAAAAUACCAUCA---CUGGCUGUAAGAUUUACAUAAAUACGGCCUGCGA-AACUAA-------AAACUGAUAAGCGUCAUAAAA--G ((.-.(((((((((.((....)).))))).............((.---(.(((((((.............))))))).).))-......-------..))))....)).........--. ( -21.52) >DroYak_CAF1 23198 108 + 1 GCCAACAGUGCGUAUGACAGUUCUUACGCAAUUAAAAUGCCAUCA---CUGGCUGUAAGAUUUACAUAAAUUCGGCCUGCGAAAACUAA-------AAACUGAUAAGCGUCAUAAAA--A ((...(((((((((.((....)).))))).............((.---(.(((((...(((((....)))))))))).).)).......-------..))))....)).........--. ( -20.20) >consensus GCCAACAGUGCGUAUGACAGUUCUUACGCAAUUAAAAUACCAUCA___CUGGCUGUAAGAUUUACAUAAAUUCGGCCUGCGA_AACUGA_______AAACUGAUAAGCGUCAUAAAA__A ((...(((((((((.((....)).))))).....................(((((...(((((....)))))))))).....................))))....))............ (-17.94 = -18.14 + 0.20)


| Location | 13,425,987 – 13,426,095 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.86 |
| Mean single sequence MFE | -26.54 |
| Consensus MFE | -17.88 |
| Energy contribution | -18.04 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.67 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.506661 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13425987 108 + 23771897 AAAUGAGGGGCA-GCAACAACA-A----------UUGGUGUGUGAACCGAAACCCGAAAGCCGAAAAGCCAGUUGGGGAAACAAUGCCAAACUCGCACAGAGCAAUGCAUUUUUCCACAA ....(((.((((-.........-.----------(((((......)))))..(((((..((......))...))))).......))))...)))(((........)))............ ( -25.50) >DroSec_CAF1 23002 106 + 1 ACAUGAGGGGCAGGCAACAACAAA----------UUGGUGUGUGAACCGAAA----AAAGCCGAAAAGCCAGUUGGGGAAACAAUGCCAAACUCGCACAGAGCAAUGCAUUUUUCCACAA ......((..((.(((.(((....----------))).))).))..))....----....((((........))))(((((.(((((....(((.....)))....)))))))))).... ( -25.40) >DroSim_CAF1 24783 106 + 1 ACAUGAGGGGCAGGCAACAACAAA----------UUGGUGUGUGAACCGAAA----AAAGCCGAAAAGCCAGUUGGGGAAACAAUGCCAAACUCGCACAGAGCAAUGCAUUUUUCCACAA ......((..((.(((.(((....----------))).))).))..))....----....((((........))))(((((.(((((....(((.....)))....)))))))))).... ( -25.40) >DroEre_CAF1 24183 104 + 1 CCAUAAGG--CA-GCAACAACA-A----------UUGGUGUGUGAACCGAAACCCGAAAGUCAAAGAGCCAGUUG--GAAACAAUGCCAAACUCGCACAGAGCACUGCAUUUUUCCACAA ......((--(.-.........-.----------(((((......)))))....(....).......))).((.(--((((.(((((....(((.....)))....)))))))))))).. ( -24.50) >DroYak_CAF1 23306 114 + 1 CCAUGAGG--CA-ACAACAACA-ACAAUUGGUUUUUGGUGUGUGAACCGAAACCCGAAAGUCAAAAAGCCAGUUG--GAAACAAUGCCAAGCUCGCAGAGAGCACUGCAUUUUUCCACAA ......((--(.-.........-......((((((.(((......)))))))))(....).......))).((.(--((((.(((((...((((.....))))...)))))))))))).. ( -31.90) >consensus ACAUGAGGGGCA_GCAACAACA_A__________UUGGUGUGUGAACCGAAACCCGAAAGCCGAAAAGCCAGUUGGGGAAACAAUGCCAAACUCGCACAGAGCAAUGCAUUUUUCCACAA .................((((.............(((((......))))).........((......))..)))).(((((.(((((....(((.....)))....)))))))))).... (-17.88 = -18.04 + 0.16)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:33:24 2006