| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,425,351 – 13,425,510 |
| Length | 159 |
| Max. P | 0.530630 |

| Location | 13,425,351 – 13,425,470 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.07 |
| Mean single sequence MFE | -30.96 |
| Consensus MFE | -23.76 |
| Energy contribution | -24.04 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.77 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.530630 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13425351 119 + 23771897 AAUAGGUCUUAAUAGAGCUAACAAUUAUUU-UUGGUAGCCACGCUUCGCGAAGCUGGGCGCAAACAAAAAAGAACGCAAACUCCGUGGCCUAUAAAUAUCGGUCUUCGGAAUUACUAAAU ..(((.(((....))).))).........(-(((((((((..((((....))))..)))......................((((.((((..........))))..))))..))))))). ( -28.10) >DroSec_CAF1 22394 120 + 1 AAUAGGUCUUAAUAGAGCUAACAAUUAUUUUUUGGUAGCCACGCUUCGCGGAGCUGGGCGCAAACAAAAAAGAACGCAAACUCCGUGGCCUAUAAAUAUCGGUCUUCGGAAUUACUAAAU ..(((.(((....))).)))..........((((((((((..((((....))))..)))......................((((.((((..........))))..))))..))))))). ( -29.00) >DroSim_CAF1 24152 120 + 1 AAUAGGUCUUAAUAGAGCUAACAAUUAUUUGUUGGUAGCCACGCUUCGCGGAGCUGGGCGCAAACAAAAAAGAACGCAAACUCCGUGGCCUAUAAAUAUCGGUCUUCGGAAUUACUAAAU ....(((.((.((((.((((((((....)))))))).(((((((...))((((.(..(((..............)))..)))))))))))))).)).)))..((....)).......... ( -30.74) >DroEre_CAF1 23454 119 + 1 AAUAGGUCUGCAUAGGCCAAACAAUUAUAU-UUGGUAGACACGCUUCGCGGAGCUGGGCGCGAAUAAAAAAGAACGCCAACUCCGUGGCCUAUAAAUAUCGGUCUUCGGAAUUACUAAAU .(((((((.((....((((((........)-)))))......))...((((((...((((..............))))..))))))))))))).........((....)).......... ( -32.04) >DroYak_CAF1 22507 119 + 1 CAUAGGUCUGCAUAGACCAAACAAUUAGAU-UUGGUAGCCACGCUUCGCGGAGCUGGGCGCAAACAAAAAAGAACGCCAACUCCGUGGCCUAUAAAUAUCGGUCUUCGGAAUUACUAAAU ..(((.((((...(((((...(((......-)))((((((((((...))((((...((((..............))))..)))))))).)))).......))))).))))....)))... ( -34.94) >consensus AAUAGGUCUUAAUAGAGCUAACAAUUAUUU_UUGGUAGCCACGCUUCGCGGAGCUGGGCGCAAACAAAAAAGAACGCAAACUCCGUGGCCUAUAAAUAUCGGUCUUCGGAAUUACUAAAU ..(((.(((....))).)))...........(((((((((..((((....))))..)))......................((((.((((..........))))..))))..)))))).. (-23.76 = -24.04 + 0.28)

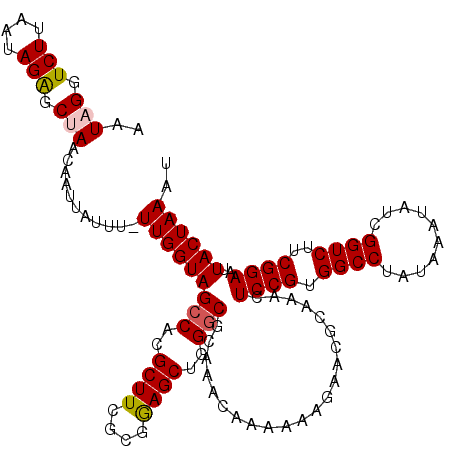

| Location | 13,425,390 – 13,425,510 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.33 |
| Mean single sequence MFE | -33.63 |
| Consensus MFE | -28.46 |
| Energy contribution | -28.74 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.85 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.519114 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13425390 120 + 23771897 ACGCUUCGCGAAGCUGGGCGCAAACAAAAAAGAACGCAAACUCCGUGGCCUAUAAAUAUCGGUCUUCGGAAUUACUAAAUCCAAUUGCAGUCCAAUUAGCAGUCGGACGCCAAUGAAGCG .((((((((...))..((((.....................((((.((((..........))))..)))).........(((.(((((..........))))).)))))))...)))))) ( -32.00) >DroSec_CAF1 22434 120 + 1 ACGCUUCGCGGAGCUGGGCGCAAACAAAAAAGAACGCAAACUCCGUGGCCUAUAAAUAUCGGUCUUCGGAAUUACUAAAUCCAAUUGCAGUCCAAUUAGCAGUCGGUCGUCAAUGAAGCG .((((((((((((.(..(((..............)))..)))))))((((............((....)).............(((((..........))))).))))......)))))) ( -30.84) >DroSim_CAF1 24192 120 + 1 ACGCUUCGCGGAGCUGGGCGCAAACAAAAAAGAACGCAAACUCCGUGGCCUAUAAAUAUCGGUCUUCGGAAUUACUAAAUCCAAUUGCAGUCCAAUUAGCAGUCGGGCGUCAAUGAAGCG .(((((((((........)))..........((.(((....((((.((((..........))))..))))..........((.(((((..........))))).)))))))...)))))) ( -33.60) >DroEre_CAF1 23493 120 + 1 ACGCUUCGCGGAGCUGGGCGCGAAUAAAAAAGAACGCCAACUCCGUGGCCUAUAAAUAUCGGUCUUCGGAAUUACUAAAUCCAAUUGCAGUCCAAUUAGCAGCCGAGCGCCAAUGAAGCG .(((((((.((.(((.(((((....................((((.((((..........))))..))))............(((((.....))))).)).))).))).))..))))))) ( -36.00) >DroYak_CAF1 22546 120 + 1 ACGCUUCGCGGAGCUGGGCGCAAACAAAAAAGAACGCCAACUCCGUGGCCUAUAAAUAUCGGUCUUCGGAAUUACUAAAUCCAAUUGCAGUCCAAUUAGCAGGCGAGCGCCAAUGAAGCG .((((((((...))..(((((...(......)..((((...((((.((((..........))))..))))...............(((..........))))))).)))))...)))))) ( -35.70) >consensus ACGCUUCGCGGAGCUGGGCGCAAACAAAAAAGAACGCAAACUCCGUGGCCUAUAAAUAUCGGUCUUCGGAAUUACUAAAUCCAAUUGCAGUCCAAUUAGCAGUCGGGCGCCAAUGAAGCG .(((((((.((.(((.(((((....................((((.((((..........))))..))))............(((((.....))))).)).))).))).))..))))))) (-28.46 = -28.74 + 0.28)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:33:22 2006