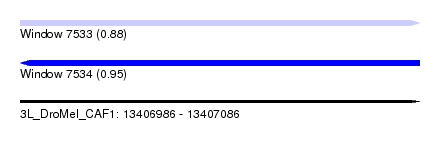
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,406,986 – 13,407,086 |
| Length | 100 |
| Max. P | 0.953656 |

| Location | 13,406,986 – 13,407,086 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.87 |
| Mean single sequence MFE | -33.45 |
| Consensus MFE | -30.42 |
| Energy contribution | -30.75 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.880314 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13406986 100 + 23771897 ---UUUCAACUGUUUCCGUGGUGUAGCGGUUAUCACAUCUGCCUAACACGCAGAAGGCCCCCGGUUCGAUCCCGGGCGGAAACAGG-UGA-UAA--ACU-UUUUUUUU------------ ---..(((.((((((((((((((.......)))))).(((((.......)))))..((((..((......)).)))))))))))).-)))-...--...-........------------ ( -34.20) >DroVir_CAF1 5601 113 + 1 AGUUCCCAACUGUUUCCGUGGUGUAGCGGUUAUCACAUCUGCCUAACACGCAGAAGGCCCCCGGUUCGAUCCCGGGCGGAAACAUG-AUA-C-A--AUUAUUUU-UUAAAUUUUCAAUG- ..........(((((((((((((.......)))))).(((((.......)))))..((((..((......)).)))))))))))..-...-.-.--........-..............- ( -28.60) >DroGri_CAF1 5402 114 + 1 UGCGUUCAACUGUUUCCGUGGUGUAGCGGUUAUCACAUCUGCCUAACACGCAGAAGGCCCCCGGUUCGAUCCCGGGCGGAAACAGA-UGA-UUU--ACU-UUUU-GCAAUUGCUUUUUUA ((((.(((.((((((((((((((.......)))))).(((((.......)))))..((((..((......)).)))))))))))).-)))-...--...-...)-)))............ ( -35.60) >DroYak_CAF1 4099 105 + 1 ---UUUCAACUGUUUCCGUGGUGUAGCGGUUAUCACAUCUGCCUAACACGCAGAAGGCCCCCGGUUCGAUCCCGGGCGGAAACAGG-UGAAGUA--CU----UUUUUGCAUCU-----UA ---(((((.((((((((((((((.......)))))).(((((.......)))))..((((..((......)).)))))))))))).-)))))..--..----...........-----.. ( -35.50) >DroAna_CAF1 3842 112 + 1 --CUUUCAGCUGUUUCCGUGGUGUAGCGGUUAUCACAUCUGCCUAACACGCAGAAGGCCCCCGGUUCGAUCCCGGGCGGAAACAGGAUGA-UUA--AUU-UUUUUGUAUAUUU-UUU-UA --...(((.((((((((((((((.......)))))).(((((.......)))))..((((..((......)).))))))))))))..)))-...--...-.............-...-.. ( -33.70) >DroPer_CAF1 4314 104 + 1 ---UUUCAACUGUUUCCGUGGUGUAGCGGUUAUCACAUCUGCCUAACACGCAGAAGGCCCCCGGUUCGAUCCCGGGCGGAAACAUG---A-UUGGGAU----UUGAUUUAUCU-----CG ---..(((..(((((((((((((.......)))))).(((((.......)))))..((((..((......)).)))))))))))))---)-..(((((----.......))))-----). ( -33.10) >consensus ___UUUCAACUGUUUCCGUGGUGUAGCGGUUAUCACAUCUGCCUAACACGCAGAAGGCCCCCGGUUCGAUCCCGGGCGGAAACAGG_UGA_UUA__AUU_UUUUUUUAAAUCU_____UA .........((((((((((((((.......)))))).(((((.......)))))..((((..((......)).))))))))))))................................... (-30.42 = -30.75 + 0.33)

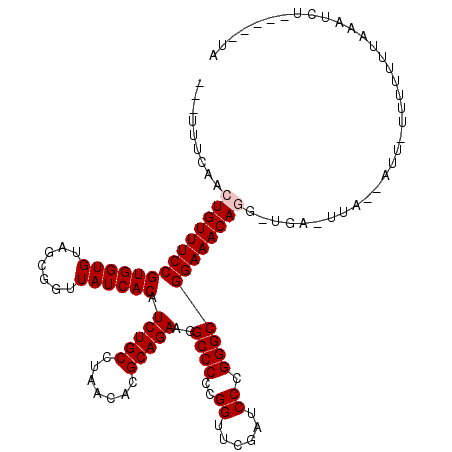

| Location | 13,406,986 – 13,407,086 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.87 |
| Mean single sequence MFE | -34.22 |
| Consensus MFE | -30.53 |
| Energy contribution | -30.87 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.43 |
| SVM RNA-class probability | 0.953656 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
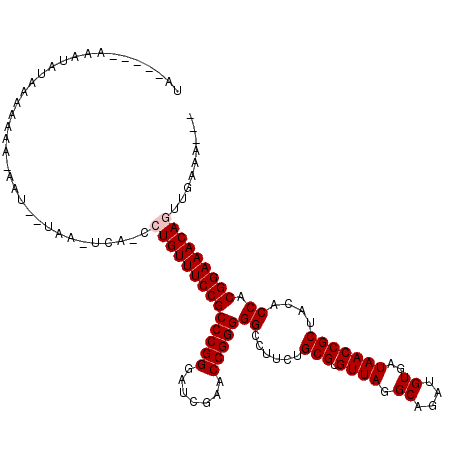
>3L_DroMel_CAF1 13406986 100 - 23771897 ------------AAAAAAAA-AGU--UUA-UCA-CCUGUUUCCGCCCGGGAUCGAACCGGGGGCCUUCUGCGUGUUAGGCAGAUGUGAUAACCGCUACACCACGGAAACAGUUGAAA--- ------------........-...--...-(((-.((((((((((((((.......)))))((......(((.((((.((....))..)))))))....)).))))))))).)))..--- ( -35.20) >DroVir_CAF1 5601 113 - 1 -CAUUGAAAAUUUAA-AAAAUAAU--U-G-UAU-CAUGUUUCCGCCCGGGAUCGAACCGGGGGCCUUCUGCGUGUUAGGCAGAUGUGAUAACCGCUACACCACGGAAACAGUUGGGAACU -..............-.......(--(-.-((.-..(((((((((((((.......)))))((......(((.((((.((....))..)))))))....)).))))))))..)).))... ( -30.50) >DroGri_CAF1 5402 114 - 1 UAAAAAAGCAAUUGC-AAAA-AGU--AAA-UCA-UCUGUUUCCGCCCGGGAUCGAACCGGGGGCCUUCUGCGUGUUAGGCAGAUGUGAUAACCGCUACACCACGGAAACAGUUGAACGCA .......((..((((-....-.))--)).-(((-.((((((((((((((.......)))))((......(((.((((.((....))..)))))))....)).))))))))).)))..)). ( -38.90) >DroYak_CAF1 4099 105 - 1 UA-----AGAUGCAAAAA----AG--UACUUCA-CCUGUUUCCGCCCGGGAUCGAACCGGGGGCCUUCUGCGUGUUAGGCAGAUGUGAUAACCGCUACACCACGGAAACAGUUGAAA--- ..-----...........----..--...((((-.((((((((((((((.......)))))((......(((.((((.((....))..)))))))....)).))))))))).)))).--- ( -35.90) >DroAna_CAF1 3842 112 - 1 UA-AAA-AAAUAUACAAAAA-AAU--UAA-UCAUCCUGUUUCCGCCCGGGAUCGAACCGGGGGCCUUCUGCGUGUUAGGCAGAUGUGAUAACCGCUACACCACGGAAACAGCUGAAAG-- ..-...-.............-...--...-(((..((((((((((((((.......)))))((......(((.((((.((....))..)))))))....)).))))))))).)))...-- ( -33.60) >DroPer_CAF1 4314 104 - 1 CG-----AGAUAAAUCAA----AUCCCAA-U---CAUGUUUCCGCCCGGGAUCGAACCGGGGGCCUUCUGCGUGUUAGGCAGAUGUGAUAACCGCUACACCACGGAAACAGUUGAAA--- .(-----(......))..----....(((-(---..(((((((((((((.......)))))((......(((.((((.((....))..)))))))....)).))))))))))))...--- ( -31.20) >consensus UA_____AAAUAUAAAAAAA_AAU__UAA_UCA_CCUGUUUCCGCCCGGGAUCGAACCGGGGGCCUUCUGCGUGUUAGGCAGAUGUGAUAACCGCUACACCACGGAAACAGUUGAAA___ ...................................((((((((((((((.......)))))((......(((.((((.((....))..)))))))....)).)))))))))......... (-30.53 = -30.87 + 0.33)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:33:12 2006