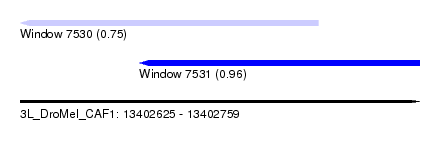
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,402,625 – 13,402,759 |
| Length | 134 |
| Max. P | 0.963296 |

| Location | 13,402,625 – 13,402,725 |
|---|---|
| Length | 100 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.75 |
| Mean single sequence MFE | -16.73 |
| Consensus MFE | -11.01 |
| Energy contribution | -11.23 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.751022 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13402625 100 - 23771897 --------------------AAUUUUCUAAACACAAUAUCUCAUAAGUAACCUUGAGAUAAUGUUUAACACCAAUUUCUAAAUUACAUCUGAUGGUCAAAGUCUAUUGGGAAAUUAAGAA --------------------.......((((((...(((((((..........))))))).))))))........((((.((((...((..((((.......))))..)).)))).)))) ( -16.70) >DroSec_CAF1 2869 114 - 1 UGACAAGAUAUCUAUCUGAAAGUUUUCUGAACACAAUAUCUCAUAAGUUACCUUGAGAUAAUGUUUAAUACCAACUUCUAAAUUUCAUCUGAAG------GUCUAUUGACAUAUUAGGAA .....((((....))))..........((((((...(((((((..........))))))).))))))...((..((((............))))------(((....)))......)).. ( -18.20) >DroSim_CAF1 6179 91 - 1 -----------------------UUUCUGAACACAAUAUCUCAUAAGUUACCUUGAGAUAAUGUUUAAUACCAACUUCUAAAUUACAUCUGAAG------AUCUAUUGACAUAUUAGGAA -----------------------.((((((......(((((((..........)))))))(((((.((((....((((............))))------...))))))))).)))))). ( -15.30) >consensus ____________________A_UUUUCUGAACACAAUAUCUCAUAAGUUACCUUGAGAUAAUGUUUAAUACCAACUUCUAAAUUACAUCUGAAG______GUCUAUUGACAUAUUAGGAA .......................(((((((..(((.(((((((..........))))))).)))..........((((............))))...................))))))) (-11.01 = -11.23 + 0.23)



| Location | 13,402,665 – 13,402,759 |
|---|---|
| Length | 94 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 72.07 |
| Mean single sequence MFE | -18.00 |
| Consensus MFE | -11.73 |
| Energy contribution | -11.73 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.65 |
| SVM decision value | 1.55 |
| SVM RNA-class probability | 0.963296 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13402665 94 - 23771897 CAAAAAUUCACGGUAAAUGGGAAUAGAUGUUGUC--------------------------AAUUUUCUAAACACAAUAUCUCAUAAGUAACCUUGAGAUAAUGUUUAACACCAAUUUCUA ...........(((....(..(((.(((...)))--------------------------.)))..)((((((...(((((((..........))))))).))))))..)))........ ( -16.30) >DroSec_CAF1 2903 119 - 1 -UAUUAUUCACGGUAAAUGGGAAUAAACAUCGGCACGUGAUGACAAGAUAUCUAUCUGAAAGUUUUCUGAACACAAUAUCUCAUAAGUUACCUUGAGAUAAUGUUUAAUACCAACUUCUA -..........((((...(..(((...(((((.....)))))...((((....))))....)))..)((((((...(((((((..........))))))).)))))).))))........ ( -21.80) >DroSim_CAF1 6213 85 - 1 -UAUUAUUCACGGUAAAUGGGAAUAAACG----------------------------------UUUCUGAACACAAUAUCUCAUAAGUUACCUUGAGAUAAUGUUUAAUACCAACUUCUA -..........((((...((((((....)----------------------------------)))))(((((...(((((((..........))))))).)))))..))))........ ( -15.90) >consensus _UAUUAUUCACGGUAAAUGGGAAUAAACGU_G_C__________________________A_UUUUCUGAACACAAUAUCUCAUAAGUUACCUUGAGAUAAUGUUUAAUACCAACUUCUA ...........((((.(((........))).....................................((((((...(((((((..........))))))).)))))).))))........ (-11.73 = -11.73 + 0.01)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:33:09 2006