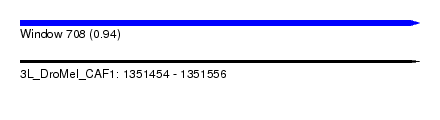
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 1,351,454 – 1,351,556 |
| Length | 102 |
| Max. P | 0.938287 |

| Location | 1,351,454 – 1,351,556 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 81.78 |
| Mean single sequence MFE | -49.03 |
| Consensus MFE | -29.06 |
| Energy contribution | -32.62 |
| Covariance contribution | 3.56 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.65 |
| Structure conservation index | 0.59 |
| SVM decision value | 1.29 |
| SVM RNA-class probability | 0.938287 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1351454 102 + 23771897 GCUCCGCCACCUGCCCUCUCUUUUCAGCCGUCUGCAACGGAAG--CAGACGCUGGGUAAAGGGAGU---------UAGGGGGUUC-GGAGGGACGGUGCAUGUCAUAAUAAUUA .(((((((.(((...(((((((((((((.((((((.......)--))))))))))..)))))))).---------..)))))..)-)))).((((.....)))).......... ( -42.20) >DroSec_CAF1 6994 100 + 1 GCUCCGCCACCUGCCCUCUCUUUUCAGCCGUCUGCGACGGAAG--CAGACACAGGGGCAAGGGAGU---------UAGG-GGUUC-GG-GGGGCGGUGCAUGUCAUAAUAAUUA ((.(((((.(((((((((((((((..(((((((((.......)--))))).....)))))))))).---------..))-)))..-))-).))))).))............... ( -43.00) >DroEre_CAF1 2823 114 + 1 GCUCCGCCACCUGCCCUCUCUUUCCUGCCGUCUGCGACGGAAGCGCAGACACAGGAGCAGAGGGGUAGGGGGCGGUAGGGGGAUCGAGGGGGGCGGUGCAUGUCAUAAUAAUUA ((.(((((.(((((((.((((((((((..(((((((.(....)))))))).)))))).)))))))))))...((((......)))).....))))).))............... ( -60.20) >DroYak_CAF1 7935 110 + 1 GCUCCGCCACCUGCCCUCUCUUUCCUGCCGUCUGCGACGGAAG--CAGACACACCGGCGGAGGGGUAGGGGGU--UCGCGGGGUUCGGGGGGGCGGUGCAUGUCAUAAUAAUUA ((.(((((.((((((((...(..((((((((((((.......)--)))))...((......))))))))..).--....)))))..)))..))))).))............... ( -50.70) >consensus GCUCCGCCACCUGCCCUCUCUUUCCAGCCGUCUGCGACGGAAG__CAGACACAGGGGCAAAGGAGU_________UAGGGGGUUC_GGGGGGGCGGUGCAUGUCAUAAUAAUUA ((.(((((.(((((((((((((((((((((((((...(....)..))))).....))))))))))...........))))))).....)))))))).))............... (-29.06 = -32.62 + 3.56)

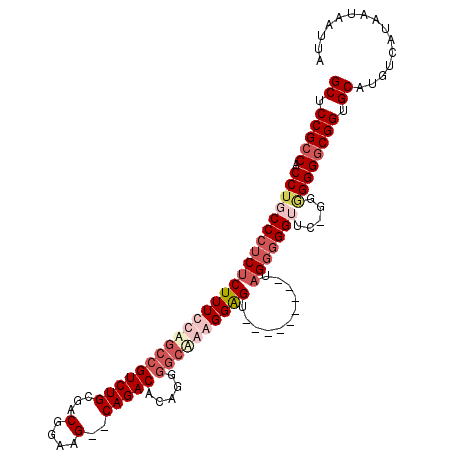
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:42:57 2006