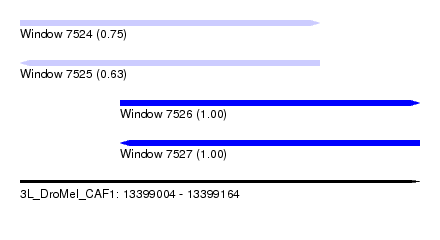
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,399,004 – 13,399,164 |
| Length | 160 |
| Max. P | 0.999859 |

| Location | 13,399,004 – 13,399,124 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.00 |
| Mean single sequence MFE | -41.86 |
| Consensus MFE | -39.24 |
| Energy contribution | -39.00 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.19 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.753549 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13399004 120 + 23771897 CAUCCUGUUUUCGGCUGGAAGAGACACAAUUGUCCAGGUAAUUUAAACCGCCGGGCAAAAUGCCGAACGUGUUAGCUAACGGCCGGUGCAUUGUGCAUUGCCCCGGCUGAUAGCCAUCGC ......((((.((((((((.((.......)).))))(((.......)))))))(((.....)))))))(((...((((.(((((((.(((........))).))))))).))))...))) ( -43.40) >DroSec_CAF1 14575 120 + 1 CAUCUUGUUUUCGACUGGAGGAGAAACAGUUGUCCAGGUAAUUUAAACCGCCGGGCAAAAUGCCGAACGUGUUAGCUAACGGCCGGUGCAUUGUGUAUUGCCCCGGCUGAUAGCCAUCGC .........((((.((((.(((.((....)).))).(((.......))).))))((.....)))))).(((...((((.(((((((.(((........))).))))))).))))...))) ( -39.00) >DroSim_CAF1 11566 120 + 1 CAUCCUGUUUUCGACUGGAGGAGAAACAGUUGUCCAGGUAAUUUAAACCGCCGGGCAAAAUGCCGAACGUGUUAGCUAACGGCCGGUGCAUUGUGUAUUGCCCCGGCUGAUAGCCAUCGC ...((((....((((((.........))))))..))))...............(((.....)))....(((...((((.(((((((.(((........))).))))))).))))...))) ( -40.60) >DroEre_CAF1 12152 120 + 1 CAUCCUGUUUUCGGUUGGAAGAGAAACAGUUGUCCAGGUAAUUCAAACCGCCGGGCAAAAUGCCGAACGCGUUAGCUAACGGCCGGUGCAUUAUGUACUGCCCCGGCUGAUAGCCAUCGC ......((((.((((((((...((.....)).))))(((.......)))))))(((.....)))))))(((...((((.(((((((.(((........))).))))))).))))...))) ( -41.40) >DroYak_CAF1 11789 120 + 1 CAUCCUGCUUUCGGUUGGUGGGGAAACAGUUGUCCAGGUAAUUCAAGCCGCCGGGCAAGAUGCCGAACGCGUUAGCUAACGGCCGGUGCAAUGUGUACUGCCCCGGCUGGUAGCCAUCGC ((((.((((..(((((((...(....)......)))(((.......))))))))))).))))......(((...((((.(((((((.(((........))).))))))).))))...))) ( -44.90) >consensus CAUCCUGUUUUCGGCUGGAGGAGAAACAGUUGUCCAGGUAAUUUAAACCGCCGGGCAAAAUGCCGAACGUGUUAGCUAACGGCCGGUGCAUUGUGUAUUGCCCCGGCUGAUAGCCAUCGC .........((((((((.........)).((((((.(((.......)))...))))))...)))))).(((...((((.(((((((.(((........))).))))))).))))...))) (-39.24 = -39.00 + -0.24)


| Location | 13,399,004 – 13,399,124 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.00 |
| Mean single sequence MFE | -40.67 |
| Consensus MFE | -34.18 |
| Energy contribution | -34.94 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.627461 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13399004 120 - 23771897 GCGAUGGCUAUCAGCCGGGGCAAUGCACAAUGCACCGGCCGUUAGCUAACACGUUCGGCAUUUUGCCCGGCGGUUUAAAUUACCUGGACAAUUGUGUCUCUUCCAGCCGAAAACAGGAUG ..(.((((((.(.(((((.(((........))).))))).).)))))).).((((((((.....)))(((((((.......))).(((((....)))))......))))......))))) ( -42.00) >DroSec_CAF1 14575 120 - 1 GCGAUGGCUAUCAGCCGGGGCAAUACACAAUGCACCGGCCGUUAGCUAACACGUUCGGCAUUUUGCCCGGCGGUUUAAAUUACCUGGACAACUGUUUCUCCUCCAGUCGAAAACAAGAUG .(((((((((.(.(((((.(((........))).))))).).))))))...(((..(((.....)))..)))...........(((((.............))))))))........... ( -36.12) >DroSim_CAF1 11566 120 - 1 GCGAUGGCUAUCAGCCGGGGCAAUACACAAUGCACCGGCCGUUAGCUAACACGUUCGGCAUUUUGCCCGGCGGUUUAAAUUACCUGGACAACUGUUUCUCCUCCAGUCGAAAACAGGAUG ..(.((((((.(.(((((.(((........))).))))).).)))))).).(((..(((.....)))..)))..........((((..(.((((.........)))).)....))))... ( -37.90) >DroEre_CAF1 12152 120 - 1 GCGAUGGCUAUCAGCCGGGGCAGUACAUAAUGCACCGGCCGUUAGCUAACGCGUUCGGCAUUUUGCCCGGCGGUUUGAAUUACCUGGACAACUGUUUCUCUUCCAACCGAAAACAGGAUG (((.((((((.(.(((((.(((........))).))))).).)))))).)))(((((((.....)))....(((.......))).))))..((((((.((........)))))))).... ( -43.20) >DroYak_CAF1 11789 120 - 1 GCGAUGGCUACCAGCCGGGGCAGUACACAUUGCACCGGCCGUUAGCUAACGCGUUCGGCAUCUUGCCCGGCGGCUUGAAUUACCUGGACAACUGUUUCCCCACCAACCGAAAGCAGGAUG (((.((((((.(.(((((.(((((....))))).))))).).)))))).)))......((((((((....(((.(((........((((....))))......))))))...)))))))) ( -44.14) >consensus GCGAUGGCUAUCAGCCGGGGCAAUACACAAUGCACCGGCCGUUAGCUAACACGUUCGGCAUUUUGCCCGGCGGUUUAAAUUACCUGGACAACUGUUUCUCCUCCAGCCGAAAACAGGAUG ..(.((((((.(.(((((.(((........))).))))).).)))))).).((((((((.....)))(((((((.......)))((((.............))))))))......))))) (-34.18 = -34.94 + 0.76)

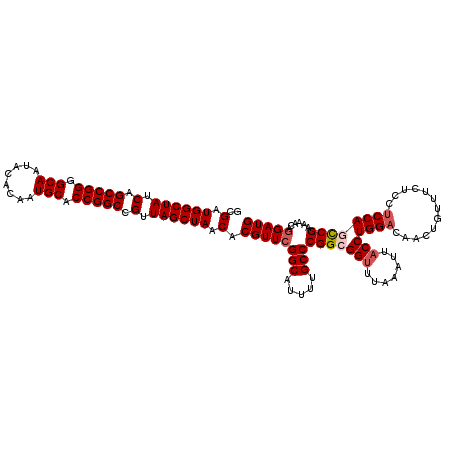
| Location | 13,399,044 – 13,399,164 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.50 |
| Mean single sequence MFE | -57.40 |
| Consensus MFE | -57.08 |
| Energy contribution | -57.08 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.06 |
| Mean z-score | -5.22 |
| Structure conservation index | 0.99 |
| SVM decision value | 4.28 |
| SVM RNA-class probability | 0.999859 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13399044 120 + 23771897 AUUUAAACCGCCGGGCAAAAUGCCGAACGUGUUAGCUAACGGCCGGUGCAUUGUGCAUUGCCCCGGCUGAUAGCCAUCGCGUUGUUCCGGCAUUUUGCCCAACUCAAAAUUGGACGCAGU .........((.(((((((((((((((((((...((((.(((((((.(((........))).))))))).))))...))))))....)))))))))))))...(((....)))..))... ( -55.80) >DroSec_CAF1 14615 120 + 1 AUUUAAACCGCCGGGCAAAAUGCCGAACGUGUUAGCUAACGGCCGGUGCAUUGUGUAUUGCCCCGGCUGAUAGCCAUCGCGUUGUUCCGGCAUUUUGCCCAGCUCAAAAUUGGACGCAGU .........((.(((((((((((((((((((...((((.(((((((.(((........))).))))))).))))...))))))....))))))))))))).))................. ( -57.70) >DroSim_CAF1 11606 120 + 1 AUUUAAACCGCCGGGCAAAAUGCCGAACGUGUUAGCUAACGGCCGGUGCAUUGUGUAUUGCCCCGGCUGAUAGCCAUCGCGUUGUUCCGGCAUUUUGCCCAGCUCAAAAUUGGACGCAGU .........((.(((((((((((((((((((...((((.(((((((.(((........))).))))))).))))...))))))....))))))))))))).))................. ( -57.70) >DroEre_CAF1 12192 120 + 1 AUUCAAACCGCCGGGCAAAAUGCCGAACGCGUUAGCUAACGGCCGGUGCAUUAUGUACUGCCCCGGCUGAUAGCCAUCGCGUUGUUCCGGCAUUUUGCCCAGCUCAAAAUUGGACGCAGU .........((.(((((((((((((((((((...((((.(((((((.(((........))).))))))).))))...))))))....))))))))))))).))................. ( -58.80) >DroYak_CAF1 11829 120 + 1 AUUCAAGCCGCCGGGCAAGAUGCCGAACGCGUUAGCUAACGGCCGGUGCAAUGUGUACUGCCCCGGCUGGUAGCCAUCGCGUUGCUCCGGCAUUCUGCCCAGCUCGAAGUUGGACGUAGU .(((.(((....(((((..((((((((((((...((((.(((((((.(((........))).))))))).))))...))))))....))))))..))))).))).)))............ ( -57.00) >consensus AUUUAAACCGCCGGGCAAAAUGCCGAACGUGUUAGCUAACGGCCGGUGCAUUGUGUAUUGCCCCGGCUGAUAGCCAUCGCGUUGUUCCGGCAUUUUGCCCAGCUCAAAAUUGGACGCAGU .........((.(((((((((((((((((((...((((.(((((((.(((........))).))))))).))))...))))))....))))))))))))).))................. (-57.08 = -57.08 + -0.00)



| Location | 13,399,044 – 13,399,164 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.50 |
| Mean single sequence MFE | -59.14 |
| Consensus MFE | -56.32 |
| Energy contribution | -56.20 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.06 |
| Mean z-score | -5.65 |
| Structure conservation index | 0.95 |
| SVM decision value | 3.52 |
| SVM RNA-class probability | 0.999338 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13399044 120 - 23771897 ACUGCGUCCAAUUUUGAGUUGGGCAAAAUGCCGGAACAACGCGAUGGCUAUCAGCCGGGGCAAUGCACAAUGCACCGGCCGUUAGCUAACACGUUCGGCAUUUUGCCCGGCGGUUUAAAU .................(((((((((((((((((....(((.(.((((((.(.(((((.(((........))).))))).).)))))).).))))))))))))))))))))......... ( -54.90) >DroSec_CAF1 14615 120 - 1 ACUGCGUCCAAUUUUGAGCUGGGCAAAAUGCCGGAACAACGCGAUGGCUAUCAGCCGGGGCAAUACACAAUGCACCGGCCGUUAGCUAACACGUUCGGCAUUUUGCCCGGCGGUUUAAAU .................(((((((((((((((((....(((.(.((((((.(.(((((.(((........))).))))).).)))))).).))))))))))))))))))))......... ( -57.40) >DroSim_CAF1 11606 120 - 1 ACUGCGUCCAAUUUUGAGCUGGGCAAAAUGCCGGAACAACGCGAUGGCUAUCAGCCGGGGCAAUACACAAUGCACCGGCCGUUAGCUAACACGUUCGGCAUUUUGCCCGGCGGUUUAAAU .................(((((((((((((((((....(((.(.((((((.(.(((((.(((........))).))))).).)))))).).))))))))))))))))))))......... ( -57.40) >DroEre_CAF1 12192 120 - 1 ACUGCGUCCAAUUUUGAGCUGGGCAAAAUGCCGGAACAACGCGAUGGCUAUCAGCCGGGGCAGUACAUAAUGCACCGGCCGUUAGCUAACGCGUUCGGCAUUUUGCCCGGCGGUUUGAAU .................(((((((((((((((((....(((((.((((((.(.(((((.(((........))).))))).).)))))).))))))))))))))))))))))......... ( -63.60) >DroYak_CAF1 11829 120 - 1 ACUACGUCCAACUUCGAGCUGGGCAGAAUGCCGGAGCAACGCGAUGGCUACCAGCCGGGGCAGUACACAUUGCACCGGCCGUUAGCUAACGCGUUCGGCAUCUUGCCCGGCGGCUUGAAU ............(((((((((((((..(((((((....(((((.((((((.(.(((((.(((((....))))).))))).).)))))).))))))))))))..)))))...)))))))). ( -62.40) >consensus ACUGCGUCCAAUUUUGAGCUGGGCAAAAUGCCGGAACAACGCGAUGGCUAUCAGCCGGGGCAAUACACAAUGCACCGGCCGUUAGCUAACACGUUCGGCAUUUUGCCCGGCGGUUUAAAU .................(((((((((((((((((....(((.(.((((((.(.(((((.(((........))).))))).).)))))).).))))))))))))))))))))......... (-56.32 = -56.20 + -0.12)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:33:05 2006