
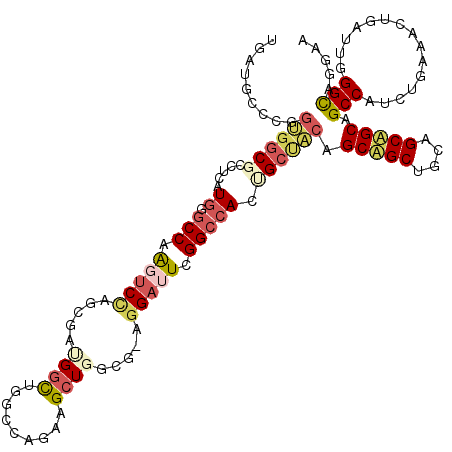
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,391,423 – 13,391,541 |
| Length | 118 |
| Max. P | 0.918708 |

| Location | 13,391,423 – 13,391,541 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 74.59 |
| Mean single sequence MFE | -49.95 |
| Consensus MFE | -21.58 |
| Energy contribution | -22.81 |
| Covariance contribution | 1.23 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.43 |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.918708 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13391423 118 + 23771897 UGAUGCUCGGUGGCGCUUCGUGUGCCAAGUCUAGCGAUGGUUGACCAGAAGCUGGCG-AGGAUUCGGCCACUGCUACGGCUGCUGCAGCAGCAGCCAUCUGAAAUUGAUUGGGCAGGAA .(((.((((.((((((.....))))))...(((((..(((....)))...)))))))-)).)))......(((((..((((((((...))))))))(((.......)))..)))))... ( -48.40) >DroGri_CAF1 4369 109 + 1 UCAAGGACACGGGCGCUUCGUGCGCCAAAUCCAA----GGCUGGU-----GCUGUGU-UUGACUCGGCAACGGCAACAGCAGCCGCAGCGGCAGCCAACUGAAACUGAUUGGGUAGAAA (((((((....(((((.....)))))...)))..----(((((.(-----((((((.-(((.((.(....)(....)))))).))))))).)))))............))))....... ( -42.90) >DroEre_CAF1 4518 118 + 1 UAAUGCCCGGUGGCGCCUCAUGGGCCAAGUCCAGCGAUGGCUGGCCAGAAGCUGGCG-AGGAUUCGGCCACAGCUACGGCUGCUGCAGCAGCCGCCAUCUGAAAUUGAUUGGGCAGGAA ...(((((((((((((..((..((((.((((((((....))).(((((...))))).-.))))).)))).((((....)))).))..)).))))))(((.......))).))))).... ( -55.50) >DroWil_CAF1 6706 115 + 1 UGAUCAUCUGCGUCGUUUCAUGGGCCAAAUCCAA----GGCUGGCGCUGUGCUGGCAUUGGUCUCGGCCACUGCUGCAGCAGCUGCAGCUGCAGCCAAUUGAAACUGAUUGGGUAGAAA ......((((((((((((((..((((....((((----.((..((.....))..)).))))....))))...((((((((.......))))))))....)))))).)))...))))).. ( -48.30) >DroYak_CAF1 4038 118 + 1 UAAUGGCGGGUGGCGCCUCAUGUGCCAAGUCCAGCGAUGGCUGGCCAGAAGCUGGCG-AGGAUUCGGCCACUGCUACGGCUGCUGCAGCAGCCGCCAUCUGAAAUUGAUUGGGCAGGAA ...((((.((((((((.....)))))).(.(((((..(((....)))...)))))).-.....)).))))((((..(((((((....)))))))(((((.......)).)))))))... ( -52.30) >DroAna_CAF1 12573 115 + 1 UGAUGUCCAGUGGCGCCUCAUGGCCCAGGUCCAGGGACGCC---CCUGAGGCUGGCG-AGGAUUCGGCCACCGCCACAGCAGCCGCGGCUGCAGCCAUAUGAAACUGGUUCGGCAGGAA .....(((.(((((.((((...(((..(((((((((....)---)))).))))))))-))).....))))).(((...(((((....)))))(((((........))))).))).))). ( -52.30) >consensus UGAUGCCCGGUGGCGCCUCAUGGGCCAAGUCCAGCGAUGGCUGGCCAGAAGCUGGCG_AGGAUUCGGCCACUGCUACAGCAGCUGCAGCAGCAGCCAUCUGAAACUGAUUGGGCAGGAA .........((((((.....((.(((.(((((.....((((.........)))).....))))).))))).)))))).(((((....))))).(((...............)))..... (-21.58 = -22.81 + 1.23)


| Location | 13,391,423 – 13,391,541 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 74.59 |
| Mean single sequence MFE | -44.97 |
| Consensus MFE | -21.06 |
| Energy contribution | -21.57 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.47 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.777433 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13391423 118 - 23771897 UUCCUGCCCAAUCAAUUUCAGAUGGCUGCUGCUGCAGCAGCCGUAGCAGUGGCCGAAUCCU-CGCCAGCUUCUGGUCAACCAUCGCUAGACUUGGCACACGAAGCGCCACCGAGCAUCA ....(((.(............((((((((((...))))))))))....((((((((....)-))...((((((((((((...((....)).))))).)).)))))))))).).)))... ( -40.40) >DroGri_CAF1 4369 109 - 1 UUUCUACCCAAUCAGUUUCAGUUGGCUGCCGCUGCGGCUGCUGUUGCCGUUGCCGAGUCAA-ACACAGC-----ACCAGCC----UUGGAUUUGGCGCACGAAGCGCCCGUGUCCUUGA ............((((..(((....)))..)))).(((((.(((((...(((......)))-...))))-----).)))))----..((((..(((((.....)))))...)))).... ( -35.60) >DroEre_CAF1 4518 118 - 1 UUCCUGCCCAAUCAAUUUCAGAUGGCGGCUGCUGCAGCAGCCGUAGCUGUGGCCGAAUCCU-CGCCAGCUUCUGGCCAGCCAUCGCUGGACUUGGCCCAUGAGGCGCCACCGGGCAUUA ....(((((..........((...((((((((....))))))))..))((((((((....)-))...(((((.(((((((((....))).).)))))...)))))))))).)))))... ( -54.00) >DroWil_CAF1 6706 115 - 1 UUUCUACCCAAUCAGUUUCAAUUGGCUGCAGCUGCAGCUGCUGCAGCAGUGGCCGAGACCAAUGCCAGCACAGCGCCAGCC----UUGGAUUUGGCCCAUGAAACGACGCAGAUGAUCA ..............((((((....((((((((.......)))))))).(.(((((((.((((.((..((.....))..)).----)))).)))))))).)))))).............. ( -43.20) >DroYak_CAF1 4038 118 - 1 UUCCUGCCCAAUCAAUUUCAGAUGGCGGCUGCUGCAGCAGCCGUAGCAGUGGCCGAAUCCU-CGCCAGCUUCUGGCCAGCCAUCGCUGGACUUGGCACAUGAGGCGCCACCCGCCAUUA ....................((((((((((((....))))))......((((((((....)-))...(((((((((((((((....))).).)))).)).))))))))))..)))))). ( -47.50) >DroAna_CAF1 12573 115 - 1 UUCCUGCCGAACCAGUUUCAUAUGGCUGCAGCCGCGGCUGCUGUGGCGGUGGCCGAAUCCU-CGCCAGCCUCAGG---GGCGUCCCUGGACCUGGGCCAUGAGGCGCCACUGGACAUCA .(((.((((..(((........)))..(((((....)))))..))))(((((((((....)-))((((..(((((---(....))))))..))))(((....))))))))))))..... ( -49.10) >consensus UUCCUGCCCAAUCAAUUUCAGAUGGCUGCUGCUGCAGCAGCCGUAGCAGUGGCCGAAUCCU_CGCCAGCUUCUGGCCAGCCAUCGCUGGACUUGGCCCAUGAAGCGCCACCGGCCAUCA ...............((((...((((..(.(((((.......))))).)..))))........(((((.......(((((....)))))..)))))....))))............... (-21.06 = -21.57 + 0.50)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:32:54 2006