
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,387,239 – 13,387,335 |
| Length | 96 |
| Max. P | 0.690236 |

| Location | 13,387,239 – 13,387,335 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 76.27 |
| Mean single sequence MFE | -32.73 |
| Consensus MFE | -23.20 |
| Energy contribution | -22.78 |
| Covariance contribution | -0.42 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.08 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.690236 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13387239 96 - 23771897 UGCCCUUGGGAUCGGCACCCAAAACGAUGGGUUGGCCGCGUUGGGUGCGCGGCAGAGUUGCGUAGAUGUUUUCUGCAAUU-AAG--CGA-----UUAUUUCGAU--- ..((....))(((((((((((......)))))..(((((((.....)))))))......))(((((.....)))))....-...--)))-----).........--- ( -32.40) >DroPse_CAF1 4668 102 - 1 UGCCCUUGGGAUCGGCUCCCAAGACAAUGGGCUGGCCGCGUUGGGUGCGCGGCAGCGUUGCAUAGAUAUUUUCUGCAAUU-UCA--CGAACCA-UAAACUUUAC-CA ....(((((((.....)))))))...((((((((.((((((.....))))))))))((((((..((.....)))))))).-...--....)))-).........-.. ( -36.90) >DroWil_CAF1 3 86 - 1 UGCCCUUGGGAUCGGCACCUAGAACAAUGGGUUGGCCUCGUUGGGUGCGGGGCAGCGUAGCGUAGAUAUUUUCUGCACAA-UAA--CAA------------------ ((((((........(((((.....(((((((....)).)))))))))))))))).....(.(((((.....))))).)..-...--...------------------ ( -27.30) >DroMoj_CAF1 79 101 - 1 UGCCCUUGGGAUCGGAGCCCAAAACAAUGGGUUGGCCACGUUGGGUGCGCGGCAGCGACGCAUAGAUAUUUUCUGCAAAA-AAAAAAAA-----UUAUCAAAAUUCA (((...((.(.(((.((((((......)))))).(((.(((.....))).)))..)))).)).(((.....))))))...-........-----............. ( -22.80) >DroAna_CAF1 8487 103 - 1 UGCCCUUGGGAUCGGCGCCCAGCACAAUGGGUUGGCCCCGUUGGGUGCGCGGCAGCGUUGCGUAGAUAUUUUCUGGAAAGGGAG--CCG-CCACAUACGGCGAU-UA .(((..((((..((((.(((.((((((((((.....)))))))...((((....))))))).((((.....))))....))).)--)))-)).))...)))...-.. ( -40.10) >DroPer_CAF1 4864 102 - 1 UGCCCUUGGGAUCGGCUCCCAAGACAAUGGGCUGGCCGCGUUGGGUGCGCGGCAGCGUUGCAUAGAUAUUUUCUGCAAUU-UCA--CGAACCA-UAAACUUUAC-CA ....(((((((.....)))))))...((((((((.((((((.....))))))))))((((((..((.....)))))))).-...--....)))-).........-.. ( -36.90) >consensus UGCCCUUGGGAUCGGCACCCAAAACAAUGGGUUGGCCGCGUUGGGUGCGCGGCAGCGUUGCAUAGAUAUUUUCUGCAAAU_UAA__CGA_____UUAACUCGAU_CA .(((((((((.......)))))......))))..(((((((.....))))))).........((((.....))))................................ (-23.20 = -22.78 + -0.42)

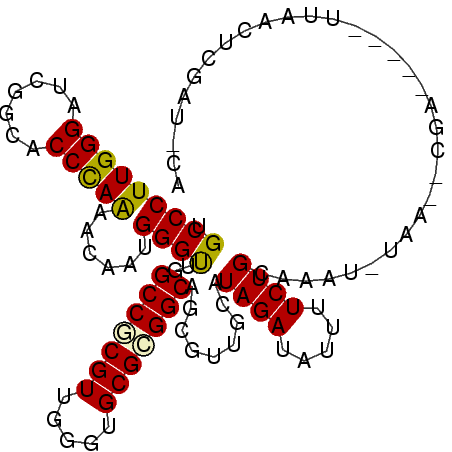
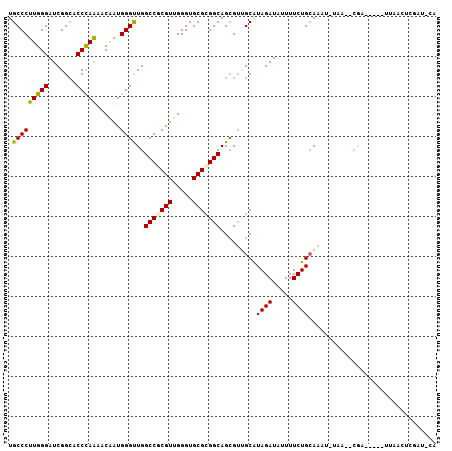
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:32:44 2006