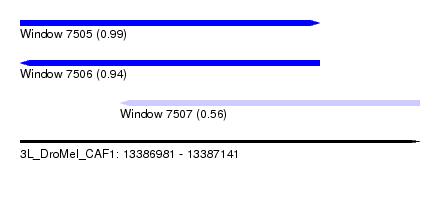
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,386,981 – 13,387,141 |
| Length | 160 |
| Max. P | 0.985791 |

| Location | 13,386,981 – 13,387,101 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.56 |
| Mean single sequence MFE | -35.99 |
| Consensus MFE | -34.15 |
| Energy contribution | -34.93 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.08 |
| Structure conservation index | 0.95 |
| SVM decision value | 2.02 |
| SVM RNA-class probability | 0.985791 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13386981 120 + 23771897 GAUACGUAGAGAGGGGGAUGAGUCAACAGGCGAAAAACGGUAAAAGCAGAAAGCAAUCGCCGGAAGCACCGUGAUUAUGCCCAUAAAUGGGCAUAGAUGCAUAUGAAAGACCUUUUUAAG ......((((((((.((.((..((....(((((......((....)).........))))).))..))))(((.(((((((((....))))))))).)))..........)))))))).. ( -34.76) >DroSim_CAF1 58543 120 + 1 GAUACGUAGAGAGGGGGAUGAGUCAACAGGCGAAAAACGGUAAAAGCAGAAAGCAAUCGCCGGAAGCGCCGUGAUUAUGCCCAUAAAAGGGCAUAGCUGCAUAUGAAAGACCUUUUUAAG .........(((((((..(...(((...((((.....((((....((.....))....))))....))))((..((((((((......))))))))..))...))).)..)))))))... ( -38.20) >DroYak_CAF1 61095 120 + 1 GACUCGUAGAGAAGGGGAUGAGUCAACAGGCGAAAAACGGUAAAAGCAGAAAGCAAUCGCCGGAAGCGCCGUGAUUAUGCCCAUAAAUGGGCAUUGCUGCAUAUGAAAGACCUUUGUAAG (((((((..........))))))).(((((((.....((((....((.....))....))))....))))((..(.(((((((....))))))).)..))..............)))... ( -35.00) >consensus GAUACGUAGAGAGGGGGAUGAGUCAACAGGCGAAAAACGGUAAAAGCAGAAAGCAAUCGCCGGAAGCGCCGUGAUUAUGCCCAUAAAUGGGCAUAGCUGCAUAUGAAAGACCUUUUUAAG .........(((((((..(...(((...((((.....((((....((.....))....))))....))))((..((((((((......))))))))..))...))).)..)))))))... (-34.15 = -34.93 + 0.78)

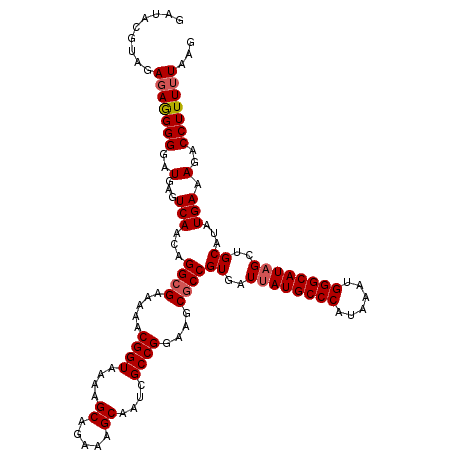
| Location | 13,386,981 – 13,387,101 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.56 |
| Mean single sequence MFE | -29.08 |
| Consensus MFE | -27.29 |
| Energy contribution | -27.40 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.27 |
| SVM RNA-class probability | 0.938827 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13386981 120 - 23771897 CUUAAAAAGGUCUUUCAUAUGCAUCUAUGCCCAUUUAUGGGCAUAAUCACGGUGCUUCCGGCGAUUGCUUUCUGCUUUUACCGUUUUUCGCCUGUUGACUCAUCCCCCUCUCUACGUAUC ........((((...((...(((((((((((((....)))))))).....)))))....(((((..((..............))...)))))))..)))).................... ( -27.54) >DroSim_CAF1 58543 120 - 1 CUUAAAAAGGUCUUUCAUAUGCAGCUAUGCCCUUUUAUGGGCAUAAUCACGGCGCUUCCGGCGAUUGCUUUCUGCUUUUACCGUUUUUCGCCUGUUGACUCAUCCCCCUCUCUACGUAUC .......(((...........(((((((((((......))))))).....((((....(((..(..((.....))..)..))).....)))).)))).........)))........... ( -29.45) >DroYak_CAF1 61095 120 - 1 CUUACAAAGGUCUUUCAUAUGCAGCAAUGCCCAUUUAUGGGCAUAAUCACGGCGCUUCCGGCGAUUGCUUUCUGCUUUUACCGUUUUUCGCCUGUUGACUCAUCCCCUUCUCUACGAGUC .....................((((((((((((....)))))))......((((....(((..(..((.....))..)..))).....)))))))))((((..............)))). ( -30.24) >consensus CUUAAAAAGGUCUUUCAUAUGCAGCUAUGCCCAUUUAUGGGCAUAAUCACGGCGCUUCCGGCGAUUGCUUUCUGCUUUUACCGUUUUUCGCCUGUUGACUCAUCCCCCUCUCUACGUAUC ........((((...((........(((((((......))))))).....((((....(((..(..((.....))..)..))).....))))))..)))).................... (-27.29 = -27.40 + 0.11)

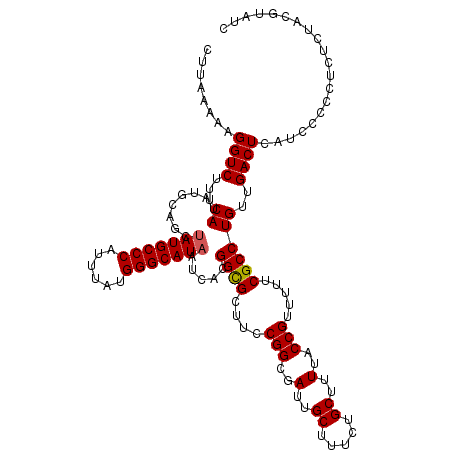
| Location | 13,387,021 – 13,387,141 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.76 |
| Mean single sequence MFE | -33.97 |
| Consensus MFE | -29.26 |
| Energy contribution | -30.04 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.564567 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13387021 120 - 23771897 GGGGUCGCUCCAACGGCUCCACACGCGCCUCCUCUUACUGCUUAAAAAGGUCUUUCAUAUGCAUCUAUGCCCAUUUAUGGGCAUAAUCACGGUGCUUCCGGCGAUUGCUUUCUGCUUUUA (((((((......))))))).....((((.(((.((........)).)))..........(((((((((((((....)))))))).....)))))....))))...((.....))..... ( -35.60) >DroSim_CAF1 58583 119 - 1 GGGGUCGCUCCAACGGCUCCUCCCCCGCCCCCUC-AACUGCUUAAAAAGGUCUUUCAUAUGCAGCUAUGCCCUUUUAUGGGCAUAAUCACGGCGCUUCCGGCGAUUGCUUUCUGCUUUUA (((((((......)))))))......(((.....-..((((...................)))).(((((((......))))))).....)))((....(((....)))....))..... ( -33.81) >DroYak_CAF1 61135 118 - 1 GGGGUCGCUCCAACUGCUCCACC-CCGCCCCCUC-AACUGCUUACAAAGGUCUUUCAUAUGCAGCAAUGCCCAUUUAUGGGCAUAAUCACGGCGCUUCCGGCGAUUGCUUUCUGCUUUUA (((((.((.......))...)))-))(((.....-..((((...................))))..(((((((....)))))))......)))((....(((....)))....))..... ( -32.51) >consensus GGGGUCGCUCCAACGGCUCCACCCCCGCCCCCUC_AACUGCUUAAAAAGGUCUUUCAUAUGCAGCUAUGCCCAUUUAUGGGCAUAAUCACGGCGCUUCCGGCGAUUGCUUUCUGCUUUUA (((((((......)))))))......(((........((((...................)))).(((((((......))))))).....)))((....(((....)))....))..... (-29.26 = -30.04 + 0.78)

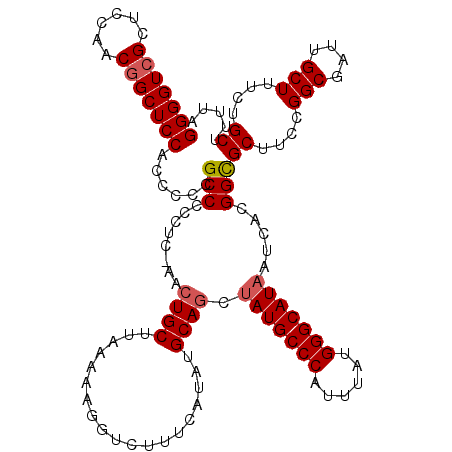
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:32:43 2006