
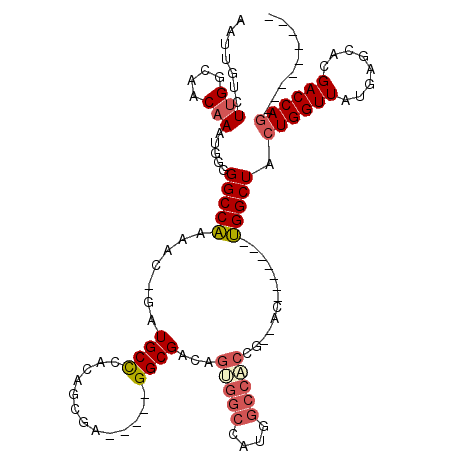
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,378,397 – 13,378,498 |
| Length | 101 |
| Max. P | 0.823495 |

| Location | 13,378,397 – 13,378,498 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.00 |
| Mean single sequence MFE | -37.55 |
| Consensus MFE | -22.03 |
| Energy contribution | -22.67 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.59 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.505097 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13378397 101 + 23771897 AAUUGUCUUGGCAACAAAUGGGGGCCAAAAC-GAUGCUCACAGCGAGGCGAGGCGACAGUGGCCAUGGCCACCG--AC-------UGGCUACUGGUUAUGAGCACGACCAG--------- .(((((.(((((..(.....)..))))).))-)))(((((.(((.((((.((.((...(((((....)))))))--.)-------).))..)).))).)))))........--------- ( -38.90) >DroPse_CAF1 28535 93 + 1 AAUUGUCUGGGCCACAAAUGGGGGCCAAAAC-GAUGCCAACUGCGA-----GGCGACAGC---CA--GCAGCCGGUGC-------UGGCUACUGGUUAUGAGCACGACCAG--------- .(((((...((((.(....)..))))...))-)))(((........-----)))...(((---((--(((.....)))-------))))).((((((........))))))--------- ( -34.80) >DroSec_CAF1 49910 110 + 1 AAUUGUCUUGGCAACAAAUGGGGGCCAAAAC-GAUGCUCACAGCGAGGCGAGGCGACAGUGGCCAUGGCCACCG--AC-------UGGCUACUGGUUAUGAGCACGACCAGGAGGAGUUC .(((((.(((((..(.....)..))))).))-)))((((..(((.((.((.(....).(((((....)))))))--.)-------).))).((((((........))))))...)))).. ( -40.70) >DroEre_CAF1 50165 95 + 1 AAUUGUCUUGGCAACAAAUGGGGGCCAAAAC-GAUGCCCACAGCGA-----GGCGACAGCGGCCAUGGCCACCG--AC-------UGGCUACUGGUUAUGAGCACGACCA---------- .(((((.(((((..(.....)..))))).))-)))((.((..((..-----(....).))(((((((((((...--..-------)))))).))))).)).)).......---------- ( -29.80) >DroYak_CAF1 52224 108 + 1 AAUUGUCUUGGCAACAAAUGGGGGCCAAAAC-GAUGCUCACAGCGA-----GGCGACAGUGGCCAUGGCCACCG--ACUUGCUACUGGCUACUGGUUAUGAGCACGACCAGGAGAG---- .(((((.(((((..(.....)..))))).))-))).(((..(((((-----(.((...(((((....)))))))--.))))))........((((((........)))))))))..---- ( -40.20) >DroAna_CAF1 49318 92 + 1 AAUUGUCUUGGCAACAAAUGUUGGCCGGAGCGGAUGCCCACGGCGA-----GGCGACUGUGGCCACCGCC-------C-------CGGCUACUGGUUAUGAGCACGACCAG--------- .......(((....)))....(((((((.((((.((.((((((((.-----..)).)))))).)))))).-------)-------))))))((((((........))))))--------- ( -40.90) >consensus AAUUGUCUUGGCAACAAAUGGGGGCCAAAAC_GAUGCCCACAGCGA_____GGCGACAGUGGCCAUGGCCACCG__AC_______UGGCUACUGGUUAUGAGCACGACCAG_________ .......(((....))).....(((((.......((((.............))))...(((((....))))).............))))).((((((........))))))......... (-22.03 = -22.67 + 0.64)


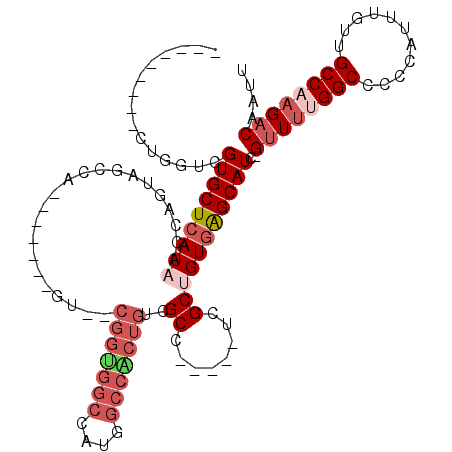
| Location | 13,378,397 – 13,378,498 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.00 |
| Mean single sequence MFE | -37.30 |
| Consensus MFE | -25.68 |
| Energy contribution | -27.48 |
| Covariance contribution | 1.81 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.823495 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13378397 101 - 23771897 ---------CUGGUCGUGCUCAUAACCAGUAGCCA-------GU--CGGUGGCCAUGGCCACUGUCGCCUCGCCUCGCUGUGAGCAUC-GUUUUGGCCCCCAUUUGUUGCCAAGACAAUU ---------......(((((((((....(..((.(-------(.--(((((((....)))))))....)).))..)..))))))))).-((((((((...........)))))))).... ( -37.00) >DroPse_CAF1 28535 93 - 1 ---------CUGGUCGUGCUCAUAACCAGUAGCCA-------GCACCGGCUGC--UG---GCUGUCGCC-----UCGCAGUUGGCAUC-GUUUUGGCCCCCAUUUGUGGCCCAGACAAUU ---------(((((..(....)..)))))((((((-------(((.....)))--))---))))..(((-----........)))...-((((.(((((......).)))).)))).... ( -35.40) >DroSec_CAF1 49910 110 - 1 GAACUCCUCCUGGUCGUGCUCAUAACCAGUAGCCA-------GU--CGGUGGCCAUGGCCACUGUCGCCUCGCCUCGCUGUGAGCAUC-GUUUUGGCCCCCAUUUGUUGCCAAGACAAUU .....((....))..(((((((((....(..((.(-------(.--(((((((....)))))))....)).))..)..))))))))).-((((((((...........)))))))).... ( -37.40) >DroEre_CAF1 50165 95 - 1 ----------UGGUCGUGCUCAUAACCAGUAGCCA-------GU--CGGUGGCCAUGGCCGCUGUCGCC-----UCGCUGUGGGCAUC-GUUUUGGCCCCCAUUUGUUGCCAAGACAAUU ----------.....(((((((....((((((...-------(.--(((((((....))))))).)..)-----).))))))))))).-((((((((...........)))))))).... ( -35.30) >DroYak_CAF1 52224 108 - 1 ----CUCUCCUGGUCGUGCUCAUAACCAGUAGCCAGUAGCAAGU--CGGUGGCCAUGGCCACUGUCGCC-----UCGCUGUGAGCAUC-GUUUUGGCCCCCAUUUGUUGCCAAGACAAUU ----.....(((((..(....)..)))))..((((.((((.((.--(((((((....)))))))....)-----).)))))).))...-((((((((...........)))))))).... ( -37.50) >DroAna_CAF1 49318 92 - 1 ---------CUGGUCGUGCUCAUAACCAGUAGCCG-------G-------GGCGGUGGCCACAGUCGCC-----UCGCCGUGGGCAUCCGCUCCGGCCAACAUUUGUUGCCAAGACAAUU ---------(((((..(....)..)))))..((((-------(-------(((((((.((((.((....-----..)).)))).)).))))))))))((((....))))........... ( -41.20) >consensus _________CUGGUCGUGCUCAUAACCAGUAGCCA_______GU__CGGUGGCCAUGGCCACUGUCGCC_____UCGCUGUGAGCAUC_GUUUUGGCCCCCAUUUGUUGCCAAGACAAUU ...............(((((((((......................(((((((....)))))))..((........)))))))))))..((((((((...........)))))))).... (-25.68 = -27.48 + 1.81)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:32:35 2006