
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,375,008 – 13,375,122 |
| Length | 114 |
| Max. P | 0.520806 |

| Location | 13,375,008 – 13,375,122 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.33 |
| Mean single sequence MFE | -33.11 |
| Consensus MFE | -21.60 |
| Energy contribution | -21.49 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.65 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.520806 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13375008 114 + 23771897 -----GGUCUUA-UUCAGUGCGGUUUUAUCUUGACUCACUGGAGACGAACACGUCCGUAUCGCUCGGUGCUUCGCACCUGACUGAAUCUUUGGUCGAUGGCUCAGUUCUAAAUUGAGCCA -----.(((((.-..(((((.((((.......)))))))))))))).....((.(((..(((.(((((((...))))).)).))).....))).)).(((((((((.....))))))))) ( -38.00) >DroPse_CAF1 23533 114 + 1 AUCUAAGUUU---UUUUGGGCAAUU---CUUUUACUCUCAGAAGACGAACACGUCCGUAUCGCUCGGUGCUUCGCACCUGACUGAAUCUUUGGCCGAUUGCUCGGUUCUAAAUUGAGCCA ..........---....((((((((---.........((((..((((....))))..........(((((...)))))...))))..........))))))))(((((......))))). ( -30.31) >DroEre_CAF1 46789 96 + 1 -----GA-----------------UU--UCUUGGCUCACUGGAGACGAACACGUCCGUAUCGCUCGGUGCUUCGCACCUGACUGAAUCUUUGGUCGAUGGCUCAGUUCUAAAUUGAGCCA -----..-----------------..--..(((((.((..(((((((....))))....(((.(((((((...))))).)).))).))).)))))))(((((((((.....))))))))) ( -30.60) >DroYak_CAF1 48389 113 + 1 -----GGUUUUAGUUCAGUGCGGGUU--UCUUAACUCACUGGAGACGAACACGUCCGUAUCGCUCGGUGCUUCGCACCUGACUGAAUCUUUGGUCGAUGGCUCAGUUCUAAAUUGAGCCA -----(..(..(((((((((((((((--....)))))....(.((((....))))).....))..(((((...)))))..)))))).))..)..)..(((((((((.....))))))))) ( -37.20) >DroAna_CAF1 45836 105 + 1 -----------A-UUUUGUGCGGAU---UCUAGACUCGAAGGAGGCGAACACGUCCGUAUCGCUCGGUGCUUCGCACCUGACUGAAUCUUUGGCCGAUUGCUCGGUUCUAAAUUGAGCCA -----------.-....((((((((---......(((....)))........)))))))).(((((((((...))))).............((((((....)))))).......)))).. ( -32.24) >DroPer_CAF1 23588 114 + 1 AUCUAAGUUU---UUUUGGGCAAUU---CUUUUACUCUCAGAAGACGAACACGUCCGUAUCGCUCGGUGCUUCGCACCUGACUGAAUCUUUGGCCGAUUGCUCGGUUCUAAAUUGAGCCA ..........---....((((((((---.........((((..((((....))))..........(((((...)))))...))))..........))))))))(((((......))))). ( -30.31) >consensus _____GGUUU___UUUUGUGCGGUU___UCUUGACUCACAGGAGACGAACACGUCCGUAUCGCUCGGUGCUUCGCACCUGACUGAAUCUUUGGCCGAUGGCUCAGUUCUAAAUUGAGCCA .............(((((((................)))))))((((....))))......(((((((((...))))).(((((((((.......)))...)))))).......)))).. (-21.60 = -21.49 + -0.11)



| Location | 13,375,008 – 13,375,122 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.33 |
| Mean single sequence MFE | -30.67 |
| Consensus MFE | -21.81 |
| Energy contribution | -21.33 |
| Covariance contribution | -0.47 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.22 |
| Structure conservation index | 0.71 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.515529 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13375008 114 - 23771897 UGGCUCAAUUUAGAACUGAGCCAUCGACCAAAGAUUCAGUCAGGUGCGAAGCACCGAGCGAUACGGACGUGUUCGUCUCCAGUGAGUCAAGAUAAAACCGCACUGAA-UAAGACC----- (((((((.........)))))))..........(((((((..(((((...)))))(((((((((....))).))).)))..(((.((.........)))))))))))-)......----- ( -31.60) >DroPse_CAF1 23533 114 - 1 UGGCUCAAUUUAGAACCGAGCAAUCGGCCAAAGAUUCAGUCAGGUGCGAAGCACCGAGCGAUACGGACGUGUUCGUCUUCUGAGAGUAAAAG---AAUUGCCCAAAA---AAACUUAGAU (((((.(((((....((((....))))....))))).)))))(((((...)))))(.(((((..(((((....)))))(((.........))---)))))).)....---.......... ( -29.70) >DroEre_CAF1 46789 96 - 1 UGGCUCAAUUUAGAACUGAGCCAUCGACCAAAGAUUCAGUCAGGUGCGAAGCACCGAGCGAUACGGACGUGUUCGUCUCCAGUGAGCCAAGA--AA-----------------UC----- (((((((.....((.((((...(((.......))))))))).(((((...)))))(((((((((....))).))).)))...)))))))...--..-----------------..----- ( -30.50) >DroYak_CAF1 48389 113 - 1 UGGCUCAAUUUAGAACUGAGCCAUCGACCAAAGAUUCAGUCAGGUGCGAAGCACCGAGCGAUACGGACGUGUUCGUCUCCAGUGAGUUAAGA--AACCCGCACUGAACUAAAACC----- (((((((.........)))))))........((.((((((..(((((...)))))(((((((((....))).))).)))..(((.(((....--))).)))))))))))......----- ( -33.10) >DroAna_CAF1 45836 105 - 1 UGGCUCAAUUUAGAACCGAGCAAUCGGCCAAAGAUUCAGUCAGGUGCGAAGCACCGAGCGAUACGGACGUGUUCGCCUCCUUCGAGUCUAGA---AUCCGCACAAAA-U----------- ((((...........((((....))))....((((((.....(((((...)))))(((((((((....))).))).)))....))))))...---....)).))...-.----------- ( -29.40) >DroPer_CAF1 23588 114 - 1 UGGCUCAAUUUAGAACCGAGCAAUCGGCCAAAGAUUCAGUCAGGUGCGAAGCACCGAGCGAUACGGACGUGUUCGUCUUCUGAGAGUAAAAG---AAUUGCCCAAAA---AAACUUAGAU (((((.(((((....((((....))))....))))).)))))(((((...)))))(.(((((..(((((....)))))(((.........))---)))))).)....---.......... ( -29.70) >consensus UGGCUCAAUUUAGAACCGAGCAAUCGACCAAAGAUUCAGUCAGGUGCGAAGCACCGAGCGAUACGGACGUGUUCGUCUCCAGUGAGUCAAGA___AACCGCACAAAA___AAACC_____ (((((.(((((.....(((....))).....))))).)))))(((((...)))))(((((((((....))).))).)))......................................... (-21.81 = -21.33 + -0.47)

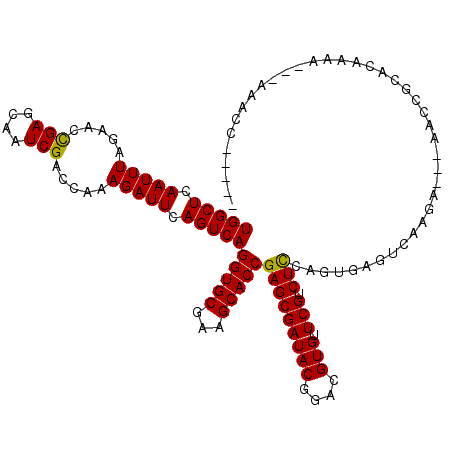

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:32:32 2006