
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,374,285 – 13,374,440 |
| Length | 155 |
| Max. P | 0.957892 |

| Location | 13,374,285 – 13,374,405 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.67 |
| Mean single sequence MFE | -31.39 |
| Consensus MFE | -24.86 |
| Energy contribution | -25.17 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.679477 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13374285 120 - 23771897 UUUUUGUACAACCCGCUGCUCUACUCGAGUGCCCUACUGUGGCCCCAAUUUUUGUUGUCCUCGGCCACGGCUCUGGGUACACCACUGACCCCCAUGACCCCCAAAUCGCCCGCCUCUGUG ......((((....((.((.........((((((..((((((((.((((....)))).....))))))))....))))))..((.((.....))))...........))..))...)))) ( -27.20) >DroPse_CAF1 22894 120 - 1 UUUCUCUAUAACCCAUUGCUGUACUCCAGUGCCCUGUUGUGGCCGCAGUUCCUGCUGUCAUCCGCCACAGCCUUGGGGACACCUUUGACACCCAUGACUCCUAAGUCCCCUGCCUCCAUA .................((.((((....))))...((((((((.(((((....))))).....))))))))...((((((........................)))))).))....... ( -29.46) >DroEre_CAF1 46070 120 - 1 UUUUUGUACAACCCGCUGCUAUACUCGAGUGCCCUACUGUGGCCCCAAUUCCUGUUGUCCUCGGCCACGGCUCUGGGCACUCCCCUGACCCCCAUGACCCCCAAAUCACCCGCCUCUGUG .............(((.((.......((((((((..((((((((.((((....)))).....))))))))....))))))))............(((........)))...))....))) ( -32.20) >DroYak_CAF1 47641 120 - 1 UUUCUGUACAACCCGCUGCUCUACUCGAGUGCCCUGCUGUGGCCCCAAUUCCUGUUGUCCUCGGCCACGGCUCUGGGUACUCCACUGACCCCCAUGACACCCAAAUCACCCGCCUCCGUG ..........................((((((((.(((((((((.((((....)))).....)))))))))...))))))))(((.((......(((........)))......)).))) ( -34.90) >DroAna_CAF1 45179 120 - 1 UUUUUGUACAACCCAUUGCUCUACUCCAGUGCUCUGCUGUGGCCCCAAUUCCUCCUGUCUUCGGCCACGGCUCUGGGCACGCCCCUGACCCCCAUGACCCCCAAGUCGCCGGCUGGCUUA .................(((........((((((.(((((((((..................)))))))))...))))))(((..((((...............))))..))).)))... ( -35.13) >DroPer_CAF1 22949 120 - 1 UUUCUCUAUAACCCAUUGCUGUACUCCAGUGCCCUGUUGUGGCCGCAGUUCCUGCUGUCAUCCGCCACAGCCUUGGGGACACCUUUGACACCCAUGACUCCUAAGUCCCCUGCCUCCAUA .................((.((((....))))...((((((((.(((((....))))).....))))))))...((((((........................)))))).))....... ( -29.46) >consensus UUUCUGUACAACCCACUGCUCUACUCCAGUGCCCUGCUGUGGCCCCAAUUCCUGCUGUCCUCGGCCACGGCUCUGGGCACACCACUGACCCCCAUGACCCCCAAAUCACCCGCCUCCGUA .................((.........((((((.(((((((((.((((....)))).....)))))))))...))))))...............((........))....))....... (-24.86 = -25.17 + 0.31)

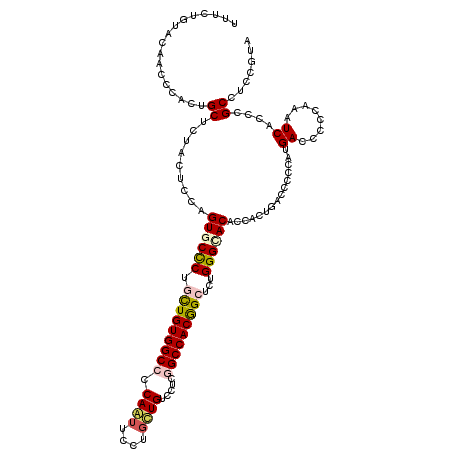
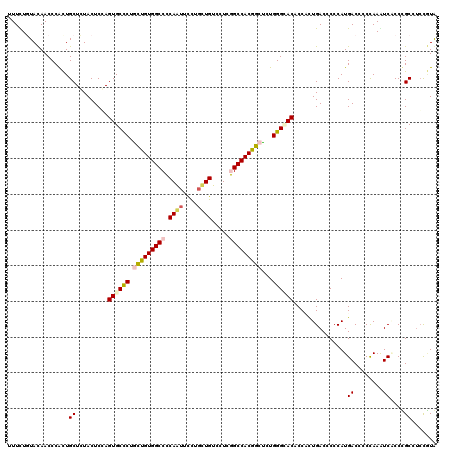
| Location | 13,374,325 – 13,374,440 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.75 |
| Mean single sequence MFE | -29.98 |
| Consensus MFE | -23.42 |
| Energy contribution | -23.48 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.48 |
| SVM RNA-class probability | 0.957892 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13374325 115 - 23771897 CUGAUUCGUUAAAUUUGCAUUU-----CAUAGAUCUGCCCUUUUUGUACAACCCGCUGCUCUACUCGAGUGCCCUACUGUGGCCCCAAUUUUUGUUGUCCUCGGCCACGGCUCUGGGUAC .......((((((...(((..(-----(...))..)))....)))).)).((((((.((((.....))))))....((((((((.((((....)))).....))))))))....)))).. ( -28.20) >DroPse_CAF1 22934 115 - 1 UCGAUCCCUUUUGAU--CAUCU---CCCGCAGAUCUGCCCUUUCUCUAUAACCCAUUGCUGUACUCCAGUGCCCUGUUGUGGCCGCAGUUCCUGCUGUCAUCCGCCACAGCCUUGGGGAC ..((((......)))--)....---...(((....))).............((((..(((((((....))))......(((((.(((((....))))).....))))))))..))))... ( -31.40) >DroEre_CAF1 46110 120 - 1 CUGACCCGUUUAAUUUGCAUUUCCCAUCAUAGAUCUGCCCUUUUUGUACAACCCGCUGCUAUACUCGAGUGCCCUACUGUGGCCCCAAUUCCUGUUGUCCUCGGCCACGGCUCUGGGCAC .......((.(((...(((..((........))..))).....))).))...................((((((..((((((((.((((....)))).....))))))))....)))))) ( -28.60) >DroYak_CAF1 47681 120 - 1 CUGAUCCGUUUUAUUCGCAUUUCCCACCGUAGAUCUGCCCUUUCUGUACAACCCGCUGCUCUACUCGAGUGCCCUGCUGUGGCCCCAAUUCCUGUUGUCCUCGGCCACGGCUCUGGGUAC ............................(((((.(.((................)).).)))))....((((((.(((((((((.((((....)))).....)))))))))...)))))) ( -33.29) >DroAna_CAF1 45219 113 - 1 -------UUAAAACCCACCUUUUAUCCCACAGAUCUGCCCUUUUUGUACAACCCAUUGCUCUACUCCAGUGCUCUGCUGUGGCCCCAAUUCCUCCUGUCUUCGGCCACGGCUCUGGGCAC -------............................(((((.............(((((........)))))....(((((((((..................)))))))))...))))). ( -26.97) >DroPer_CAF1 22989 115 - 1 UCGAUCCCUUUUGAU--CAUCU---CCCGCAGAUCUGCCCUUUCUCUAUAACCCAUUGCUGUACUCCAGUGCCCUGUUGUGGCCGCAGUUCCUGCUGUCAUCCGCCACAGCCUUGGGGAC ..((((......)))--)....---...(((....))).............((((..(((((((....))))......(((((.(((((....))))).....))))))))..))))... ( -31.40) >consensus CUGAUCCGUUUAAUU_GCAUUU___CCCACAGAUCUGCCCUUUCUGUACAACCCACUGCUCUACUCCAGUGCCCUGCUGUGGCCCCAAUUCCUGCUGUCCUCGGCCACGGCUCUGGGCAC ............................(((((.........))))).....................((((((.(((((((((.((((....)))).....)))))))))...)))))) (-23.42 = -23.48 + 0.06)


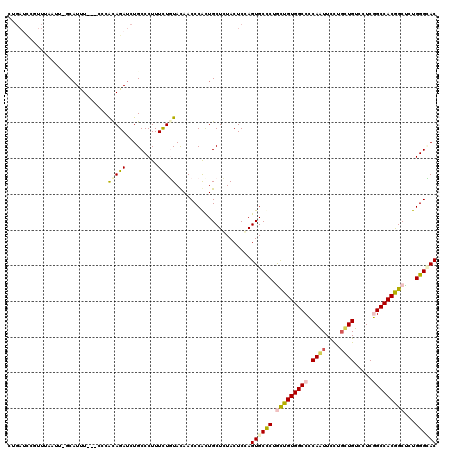
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:32:30 2006