| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,367,341 – 13,367,436 |
| Length | 95 |
| Max. P | 0.591142 |

| Location | 13,367,341 – 13,367,436 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 77.93 |
| Mean single sequence MFE | -27.37 |
| Consensus MFE | -16.73 |
| Energy contribution | -17.65 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.591142 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
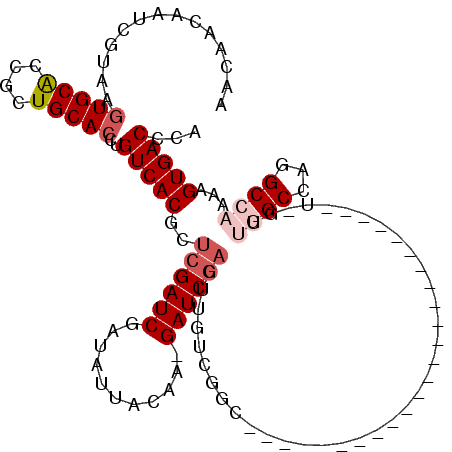
>3L_DroMel_CAF1 13367341 95 - 23771897 AACAACAGUCGUAAGUGCGCCGCUGCACCUGUCACGCUCGAUCGAUAUUACAACGAUCGACUUGUCGGC------------------------UCGUUGGCAUGGCCAAAAGUGACCCA .......((((....((.((((.(((..(.(((......))).)......((((((((((....)))).------------------------)))))))))))))))....))))... ( -26.10) >DroPse_CAF1 15299 86 - 1 AACAACAAUUGUAAGUGCACCUCUGCACCUGUCACGCUCGAUCGAUAUUACAA-GAUCGACUUGUCG--------------------------------GCCAGGCCAAAAGUGACCCA ..............(((((....)))))..(((((..((((((..........-))))))......(--------------------------------((...)))....)))))... ( -21.90) >DroEre_CAF1 39315 95 - 1 AACAACAGUCGCAAGUGCGCCGCUGCACCUGUCACGUUCGAUCGAUAUUACAACGAUCGACUUGCCGGC------------------------UUGUUGGCCAGGCCAAAAGUGACCCA .....(((((((....)))..)))).....(((((..(((((((.........)))))))...((((((------------------------(....)))).))).....)))))... ( -31.40) >DroYak_CAF1 40416 119 - 1 AACAACAGUCGUAAGUGCGCCGCUGCACCUGUCACGCUCGAUCGAUAUUACAACGAUCGACUUGUCGGCUUCUUGGCUCGUUGGCUUGUUCGCUUGUUGGCCAGGCCAAAAGUGACCCA ..(((((((((...(((((....)))))(((.((.(.(((((((.........)))))))).)).))).....))))).))))......((((((.(((((...))))))))))).... ( -35.90) >DroAna_CAF1 38413 103 - 1 AACAACAAUCGCAAAUGCACCACUGCACCUGUCACGCUCGAUCGAUAUUACAA-GAUC----UUUUGGCCUUUUG-----UUGC------CACUGCCUGGCCUGGCCAAAAGUGACCCA ...............((((....))))...(((((....((((..........-))))----((((((((.....-----..((------((.....))))..)))))))))))))... ( -27.00) >DroPer_CAF1 15319 86 - 1 AACAACAAUUGUAAGUGCACCUCUGCACCUGUCACGCUCGAUCGAUAUUACAA-GAUCGACUUGUCG--------------------------------GCCAGGCCAAAAGUGACCCA ..............(((((....)))))..(((((..((((((..........-))))))......(--------------------------------((...)))....)))))... ( -21.90) >consensus AACAACAAUCGUAAGUGCACCGCUGCACCUGUCACGCUCGAUCGAUAUUACAA_GAUCGACUUGUCGGC________________________U_GUUGGCCAGGCCAAAAGUGACCCA ..............(((((....)))))..(((((..((((((...........)))))).....................................((((...))))...)))))... (-16.73 = -17.65 + 0.92)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:32:23 2006