
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,366,575 – 13,366,734 |
| Length | 159 |
| Max. P | 0.920269 |

| Location | 13,366,575 – 13,366,676 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 88.42 |
| Mean single sequence MFE | -38.02 |
| Consensus MFE | -31.33 |
| Energy contribution | -32.17 |
| Covariance contribution | 0.83 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.663569 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
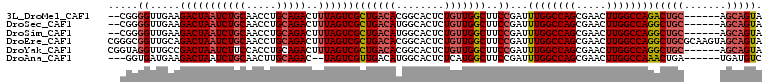
>3L_DroMel_CAF1 13366575 101 + 23771897 --CGGGGUUGAAGACUAAUCUGCAACCUGCAGACUUUAGUCGCUGACACGGCACUCUGUUGGCUUCCGAUUUGGCCAGCGAACUUGGCCAGACUGC------AGCAGUA --((((((....(((((((((((.....)))))..))))))((((...))))))))))...(((..((.(((((((((.....))))))))).)).------))).... ( -37.10) >DroSec_CAF1 38178 101 + 1 --CGGGGUUGAAGACUAAUCUGCAACCUGCAGACUUUAGUCGCUGACAUGGCACUCUGUUGGCUUCCGAUUUGGCCAGCGAACUUGGCCAGGCUGC------AGCAGUA --..........(((((((((((.....)))))..))))))((((...((((....((((((((........))))))))......)))).....)------))).... ( -37.30) >DroSim_CAF1 38076 101 + 1 --CGGGGUUGAAGACUAAUCUGCAACCUGCAGACUUUAGUCGCUGACAUGGCACUCUGUUGGCUUCCGAUUUGGCCAGCGAACUUGGCCAGGCUGC------AGCAGUA --..........(((((((((((.....)))))..))))))((((...((((....((((((((........))))))))......)))).....)------))).... ( -37.30) >DroEre_CAF1 38574 109 + 1 CGGGCGGUUGCAGACUAAUCUGCAACCUGCAGACUUUAGUCGCUGACACGGCACUCUGUUGGCUUCCGAUUUGGCCAGCGAACUUGGCCAGGCUGCGCAAGUAGCAGUA ((((((((((((((....))))))))).)).(((....)))((..(((........)))..))..)))..((((((((.....))))))))(((((....))))).... ( -50.60) >DroYak_CAF1 39641 103 + 1 CGGUAGGUUGCCGACUAAUCUUCCACCUGCAGACUUUAGUCGCUGACACGGCACUCUGUUGGCUUCCGAUUUGGCCAGCGAACUUGGCCAGGCUGC------AGCAGUA .((.((((((.....)))))).))..((((((.........((..(((........)))..))......(((((((((.....)))))))))))))------))..... ( -33.00) >DroAna_CAF1 37960 98 + 1 ---GGUGAUGAAGACUAAUCUGCAACUUGCAGAC--UAGUCGUUGACAUGGCACUCUCAUGGCUUCCGAUUUGGCCAGCGAACUUGGCCAAACUGA------UGAUGUC ---(((.((((.(((((.(((((.....))))).--)))))(((.....)))....)))).)))..((.(((((((((.....))))))))).)).------....... ( -32.80) >consensus __CGGGGUUGAAGACUAAUCUGCAACCUGCAGACUUUAGUCGCUGACACGGCACUCUGUUGGCUUCCGAUUUGGCCAGCGAACUUGGCCAGGCUGC______AGCAGUA .....((.....(((((((((((.....)))))..))))))(((((((........)))))))..))...((((((((.....))))))))(((((.......))))). (-31.33 = -32.17 + 0.83)

| Location | 13,366,612 – 13,366,706 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 85.11 |
| Mean single sequence MFE | -35.82 |
| Consensus MFE | -26.57 |
| Energy contribution | -27.43 |
| Covariance contribution | 0.86 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.652277 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13366612 94 + 23771897 UCGCUGACACGGCACUCUGUUGGCUUCCGAUUUGGCCAGCGAACUUGGCCAGACUGC------AGCAGUAUUCGUUCUCUUGGCCAAGUCUAGUUAGCUG ..((((((..........(((((((........)))))))((.((((((((((..((------..........))..)).))))))))))..)))))).. ( -35.80) >DroSec_CAF1 38215 94 + 1 UCGCUGACAUGGCACUCUGUUGGCUUCCGAUUUGGCCAGCGAACUUGGCCAGGCUGC------AGCAGUAUUCGUUCUGUUGGCCAAGUUUAGUUAGCUG ..((((((..........(((((((........)))))))(((((((((((......------.((((........))))))))))))))).)))))).. ( -38.81) >DroSim_CAF1 38113 94 + 1 UCGCUGACAUGGCACUCUGUUGGCUUCCGAUUUGGCCAGCGAACUUGGCCAGGCUGC------AGCAGUAUUCGUUCUGUUGGCCAAGUUUAGUUAGCCG ..((((((..........(((((((........)))))))(((((((((((......------.((((........))))))))))))))).)))))).. ( -38.41) >DroEre_CAF1 38613 97 + 1 UCGCUGACACGGCACUCUGUUGGCUUCCGAUUUGGCCAGCGAACUUGGCCAGGCUGCGCAAGUAGCAGUAUUCGUUCUCUUGGCCAAGUCUGAUUAG--- ..((..(((........)))..)).((.(((((((((((.((((...((...(((((....))))).))....)))).).)))))))))).))....--- ( -40.10) >DroYak_CAF1 39680 91 + 1 UCGCUGACACGGCACUCUGUUGGCUUCCGAUUUGGCCAGCGAACUUGGCCAGGCUGC------AGCAGUAUGCGUUCUCUUGGCCACGUCUGAUUAG--- ...((((((.(((....((((((((........))))))))....((((((((.(((------(......))))....)))))))).))))).))))--- ( -31.50) >DroAna_CAF1 37994 91 + 1 UCGUUGACAUGGCACUCUCAUGGCUUCCGAUUUGGCCAGCGAACUUGGCCAAACUGA------UGAUGUCUUGG-GCACUGAGUCAUGUCUGA--AGCAG ..(((((((((((.(.((((.(((.((((.(((((((((.....))))))))).)).------.)).))).)))-)....).))))))))...--))).. ( -30.30) >consensus UCGCUGACACGGCACUCUGUUGGCUUCCGAUUUGGCCAGCGAACUUGGCCAGGCUGC______AGCAGUAUUCGUUCUCUUGGCCAAGUCUAAUUAGC_G ..(((.....)))....((((((((........)))))))).(((((((((((((((.......)))))..........))))))))))........... (-26.57 = -27.43 + 0.86)


| Location | 13,366,642 – 13,366,734 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 77.29 |
| Mean single sequence MFE | -30.89 |
| Consensus MFE | -17.82 |
| Energy contribution | -17.21 |
| Covariance contribution | -0.61 |
| Combinations/Pair | 1.36 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.58 |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.920269 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
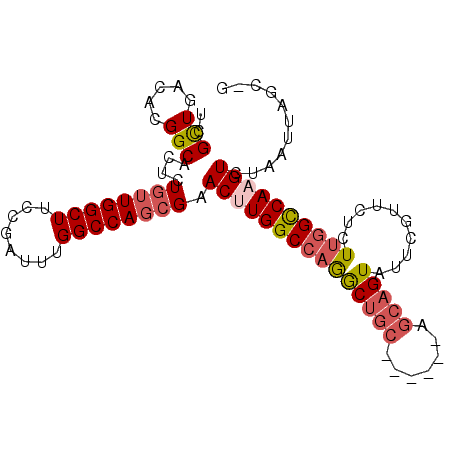
>3L_DroMel_CAF1 13366642 92 + 23771897 UUUGGCCAGCGAACUUGGCCAGACUGC------AGCAGUA-UUCGUUCUCUUGGCC-------AAGUCUAG--UUAGCUGGUCACUGGUCAA--UGAAAUCUCUACCUGG- .((((((((.(((((((((((((..((------.......-...))..)).)))))-------))))((((--....)))))).))))))))--................- ( -31.90) >DroPse_CAF1 14747 105 + 1 UUUGGCCAGCGAACUUGGCCAAACUGU------UGUUGCUUUUUGUUCCAUCAGUCUCGCCCUCAGUCCAGCCUUAGCUGACGUCUGAUUCGUUUCCAAUCUCAACCUCAA .((((..((((((...(((...((((.------((..((.....))..)).))))...))).((((.(((((....))))..).)))))))))).))))............ ( -27.00) >DroSec_CAF1 38245 92 + 1 UUUGGCCAGCGAACUUGGCCAGGCUGC------AGCAGUA-UUCGUUCUGUUGGCC-------AAGUUUAG--UUAGCUGGUCACUGGUCAA--UGAAAUCUCUACCUGG- ..(((((((((((((((((((......------.((((..-......)))))))))-------))))))..--...))))))))(.(((...--.((....)).))).).- ( -32.21) >DroSim_CAF1 38143 92 + 1 UUUGGCCAGCGAACUUGGCCAGGCUGC------AGCAGUA-UUCGUUCUGUUGGCC-------AAGUUUAG--UUAGCCGGUCACUGGUCAA--UGAAAUCUCUACCUGU- .(((((.((((((((((((((......------.((((..-......)))))))))-------)))))).)--)).)))))..((.(((...--.((....)).))).))- ( -28.21) >DroEre_CAF1 38643 94 + 1 UUUGGCCAGCGAACUUGGCCAGGCUGCGCAAGUAGCAGUA-UUCGUUCUCUUGGCC-------AAGUCUGA--UUAG----GCACUGGCCAG--UGAAAUCUCUACCUGG- .((((((((.(.(((((((((((((((....)))))((..-......)).))))))-------))))....--....----.).))))))))--................- ( -36.50) >DroYak_CAF1 39710 88 + 1 UUUGGCCAGCGAACUUGGCCAGGCUGC------AGCAGUA-UGCGUUCUCUUGGCC-------ACGUCUGA--UUAG----GCACUGGUCAA--UGAAAUCUCUACCUGG- .((((((((......((((((((.(((------(......-))))....)))))))-------).((((..--..))----)).))))))))--................- ( -29.50) >consensus UUUGGCCAGCGAACUUGGCCAGGCUGC______AGCAGUA_UUCGUUCUCUUGGCC_______AAGUCUAG__UUAGCUGGUCACUGGUCAA__UGAAAUCUCUACCUGG_ .((((((((.....))))))))(((((.......)))))......(((..((((((..............................))))))...)))............. (-17.82 = -17.21 + -0.61)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:32:22 2006