| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
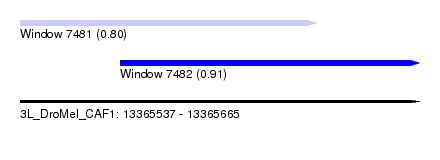
| Location | 13,365,537 – 13,365,665 |
| Length | 128 |
| Max. P | 0.908058 |

| Location | 13,365,537 – 13,365,632 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 81.94 |
| Mean single sequence MFE | -37.82 |
| Consensus MFE | -27.05 |
| Energy contribution | -26.80 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.802636 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13365537 95 + 23771897 AAAGCCGCUCGGCAA-----AAGUCCUGGAUGC--GUGGCAUGUGGCUUGUGCCACAAACGAGCCGCAUGA-U-CGAACUGGGCCUAAGA----ACCGGGAUCGGAAU----------- ....((((((((...-----.((.((..(...(--(...((((((((((((.......)))))))))))).-.-))..)..)).))....----.)))))..)))...----------- ( -38.20) >DroSec_CAF1 37142 95 + 1 AAAGCCACUCGCCAA-----AAGUCCUGGAUGC--GUGGCAUGUGGCUUGUGCCACAAACGAGCCGCAGGA-U-CGAACUGGGCCUAAGA----ACCGGGAUCGGAAU----------- ...(((((.(((((.-----......))).)).--))))).((((((((((.......))))))))))...-(-(((.((((........----.))))..))))...----------- ( -36.70) >DroSim_CAF1 37040 95 + 1 AAAGCCGCUCGGCCA-----AAGUCCUGGAUGC--GUGGCAUGUGGCUUGUGCCACAAACGAGCCGCAGGA-U-CGAACUGGGCCUAAGA----ACCGGGAUCGGAAU----------- ...((.(((((((((-----..(((...)))..--.)))).((((((....))))))..))))).))....-(-(((.((((........----.))))..))))...----------- ( -37.50) >DroEre_CAF1 37538 95 + 1 AAAGCCGCUCAGCAA-----AAGUCCUGGAUGC--GUGGCAUGUGGCUUGUGCCACAAACGAGCCGCAGGA-U-CGAACUGGGCCUAAGA----ACCGGGAUCGGAAU----------- ...(((((.((.((.-----......))..)).--))))).((((((((((.......))))))))))...-(-(((.((((........----.))))..))))...----------- ( -33.40) >DroYak_CAF1 38565 106 + 1 AAAGCCGCUCAGCAA-----AAGUCCUGGAUGC--GUGGCAUGUGGCUUGUGCCACAAACGAGCCGCAGGA-U-CGAACUGUGCCUAAGA----ACCGGGAUCCGAAUCGGGAUCGGGA ....(((.((.....-----..(((((((...(--..((((((((((((((.......)))))))))((..-.-....)))))))...).----.)))))))(((...))))).))).. ( -39.00) >DroPer_CAF1 13292 105 + 1 AAGGC---UUGCCAAAUGAGAGGUCCUUGUUCCCGGUGGCAUGUGGCUUGUGCCACAAACGAGCCGCAAUAAGCGGCACCAGGUCCAGGACAGCACCGGGACUGAGAC----------- ..(((---((..(....)..)))))(((..((((((((.(.((((((....)))))).....(((((.....))))).....(((...))).)))))))))..)))..----------- ( -42.10) >consensus AAAGCCGCUCGGCAA_____AAGUCCUGGAUGC__GUGGCAUGUGGCUUGUGCCACAAACGAGCCGCAGGA_U_CGAACUGGGCCUAAGA____ACCGGGAUCGGAAU___________ ......................(((((((......(((((.((((((....)))))).....)))))...........((.......))......)))))))................. (-27.05 = -26.80 + -0.25)



| Location | 13,365,569 – 13,365,665 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.79 |
| Mean single sequence MFE | -38.64 |
| Consensus MFE | -23.96 |
| Energy contribution | -23.97 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.28 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.62 |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.908058 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13365569 96 + 23771897 CAUGUGGCUUGUGCCACAAACGAGCCGCAUGA-U-CGAACUGGGCCUAAGA----ACCGGGAUCGGAAU------------------CGGGAUCGGGAAUCGGUACUUGGGAAACAGCCC ((((((((((((.......)))))))))))).-.-....(((..((((((.----(((((..((.((..------------------.....)).))..))))).))))))...)))... ( -40.40) >DroSec_CAF1 37174 96 + 1 CAUGUGGCUUGUGCCACAAACGAGCCGCAGGA-U-CGAACUGGGCCUAAGA----ACCGGGAUCGGAAU------------------CGGGAUCGGGAAUCGGUACUCGGGAAGAAGCCC ..((((((((((.......))))))))))...-.-......((((......----.((((((((((..(------------------((....)))...))))).)))))......)))) ( -35.22) >DroSim_CAF1 37072 96 + 1 CAUGUGGCUUGUGCCACAAACGAGCCGCAGGA-U-CGAACUGGGCCUAAGA----ACCGGGAUCGGAAU------------------CGGGAUCGGGAAUCGGUACUCGGGAAGAAGCCC ..((((((((((.......))))))))))...-.-......((((......----.((((((((((..(------------------((....)))...))))).)))))......)))) ( -35.22) >DroEre_CAF1 37570 102 + 1 CAUGUGGCUUGUGCCACAAACGAGCCGCAGGA-U-CGAACUGGGCCUAAGA----ACCGGGAUCGGAAU------------CGGGAUCGGGAUCGGGAAUCGGUACUUGGGAAGAAGCCC ..((((((((((.......))))))))))...-.-....((...((((((.----(((((..((((..(------------((....)))..))))...))))).)))))).))...... ( -37.90) >DroYak_CAF1 38597 114 + 1 CAUGUGGCUUGUGCCACAAACGAGCCGCAGGA-U-CGAACUGUGCCUAAGA----ACCGGGAUCCGAAUCGGGAUCGGGAUCGGGAUCGGGAUCGGGAAUCGGUACUUGGAAAGCAGCCC ..((((((((((.......))))))))))...-.-....((((..(((((.----(((((..(((((.((((..(((....)))..))))..)))))..))))).)))))...))))... ( -46.10) >DroPer_CAF1 13328 88 + 1 CAUGUGGCUUGUGCCACAAACGAGCCGCAAUAAGCGGCACCAGGUCCAGGACAGCACCGGGACUGAGAC------------------CGGGAUCGAGA-------CCCGAGAA------- ..((((((....))))))..((((((((.....))))).((.(((((((..(.......)..))).)))------------------).)).)))...-------........------- ( -37.00) >consensus CAUGUGGCUUGUGCCACAAACGAGCCGCAGGA_U_CGAACUGGGCCUAAGA____ACCGGGAUCGGAAU__________________CGGGAUCGGGAAUCGGUACUCGGGAAGAAGCCC ..((((((((((.......))))))))))..........((((.(((...........))).)))).....................((((((((.....)))).))))........... (-23.96 = -23.97 + 0.01)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:32:19 2006