
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,362,909 – 13,363,016 |
| Length | 107 |
| Max. P | 0.620631 |

| Location | 13,362,909 – 13,363,016 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 80.86 |
| Mean single sequence MFE | -30.32 |
| Consensus MFE | -16.26 |
| Energy contribution | -16.48 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.620631 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13362909 107 + 23771897 UUCAUACCUUUAUUAAGAAACAGAAACUCACCUGGCGAUCACUGCCACCGGUAGCAAAUUUGUCGCCAUCGCGACUGAAUGCCACCGCAUUCACCGCAUCAGUGUGG ..(((((.........................(((((((..(((((...))))).......)))))))..(((..(((((((....))))))).)))....))))). ( -29.40) >DroSec_CAF1 34403 107 + 1 UUCAUACCUUCAUAAAGAAACAGAAACUCACCUGGCGAUCACUACCACCGGUAGCAAACUUGUCGCCAUCGUGACUGAAUGCCACCGCAUUCACCGCAUCAGUGUGG ...........................((((.(((((((..(((((...))))).......)))))))..)))).(((((((....)))))))(((((....))))) ( -28.90) >DroSim_CAF1 34328 107 + 1 UCCAUACCUUCAUAAAGAAACAGAAACUCACCUGGCGAUCACUACCACCGGUAGCAAACUUGUCGCCAUCGUGACUGAAUGCCACCGCAUUUACCGCAUCAGUGUGG .((((((....................((((.(((((((..(((((...))))).......)))))))..)))).(((((((....)))))))........)))))) ( -28.40) >DroEre_CAF1 34928 106 + 1 UGCGCACA-UUAUCAAGAGACAGAAACUCACCUGACGAUCACUACCACCGGUGGCAAACUUGUCGCCAUCGCGACUGAAUGCCACCGCAUUGACCGCAUCAGUGUGG ..(((((.-.......(((.......)))...((.((.(((.......((((((((.....(((((....)))))....))))))))...))).))))...))))). ( -32.70) >DroYak_CAF1 35807 99 + 1 AA-------CAAUCAAGAAACA-AAACUCACCUGACGAUCACUGCCACCGGUGGCAAACUUGUCGCCAUCGCGACUGAAUGCCACCGCAUUGACCGCAUCAGUGUGG ..-------.............-.....(((((((((.(((.(((....(((((((.....(((((....)))))....)))))))))).))).))..)))).))). ( -33.70) >DroPer_CAF1 9688 103 + 1 GCCAU----UGGAUGAGCCGCCCCAACUCACCUGGCGAUCGUUGCCACCAGUGGCAAACUUCUCUCCAUCCUGGGAAAAGCCCACAGCAUUUACAGCCGCCGUAUGG (((((----(((.((((.........))))..((((((...))))))))))))))..........((((.(((((.....))))..((.......))....).)))) ( -28.80) >consensus UCCAUACC_UCAUAAAGAAACAGAAACUCACCUGGCGAUCACUACCACCGGUAGCAAACUUGUCGCCAUCGCGACUGAAUGCCACCGCAUUCACCGCAUCAGUGUGG ............................(((((((.......(((.....)))((......(((((....)))))(((((((....)))))))..)).)))).))). (-16.26 = -16.48 + 0.23)

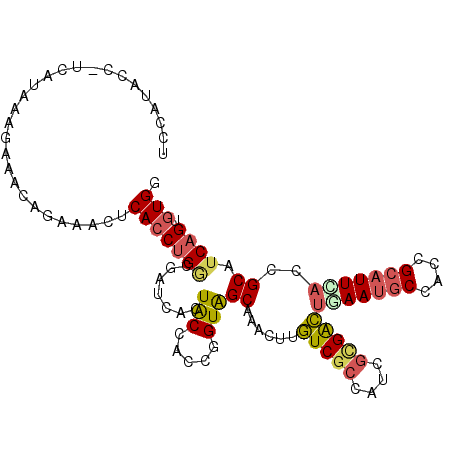
| Location | 13,362,909 – 13,363,016 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 80.86 |
| Mean single sequence MFE | -36.47 |
| Consensus MFE | -24.93 |
| Energy contribution | -23.38 |
| Covariance contribution | -1.55 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.592917 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13362909 107 - 23771897 CCACACUGAUGCGGUGAAUGCGGUGGCAUUCAGUCGCGAUGGCGACAAAUUUGCUACCGGUGGCAGUGAUCGCCAGGUGAGUUUCUGUUUCUUAAUAAAGGUAUGAA ...((((..((((((((((((....))))))).))))).((((((((...((((((....)))))))).))))))))))...(((....((((....))))...))) ( -34.80) >DroSec_CAF1 34403 107 - 1 CCACACUGAUGCGGUGAAUGCGGUGGCAUUCAGUCACGAUGGCGACAAGUUUGCUACCGGUGGUAGUGAUCGCCAGGUGAGUUUCUGUUUCUUUAUGAAGGUAUGAA .((.(((...(((((((((((....))))))).((((..((((((.....(..(((((...)))))..))))))).))))....))))(((.....)))))).)).. ( -33.90) >DroSim_CAF1 34328 107 - 1 CCACACUGAUGCGGUAAAUGCGGUGGCAUUCAGUCACGAUGGCGACAAGUUUGCUACCGGUGGUAGUGAUCGCCAGGUGAGUUUCUGUUUCUUUAUGAAGGUAUGGA (((.(((...((((..(((((....)))))...((((..((((((.....(..(((((...)))))..))))))).))))....))))(((.....)))))).))). ( -31.80) >DroEre_CAF1 34928 106 - 1 CCACACUGAUGCGGUCAAUGCGGUGGCAUUCAGUCGCGAUGGCGACAAGUUUGCCACCGGUGGUAGUGAUCGUCAGGUGAGUUUCUGUCUCUUGAUAA-UGUGCGCA .(((.(((..(((((((.((((((((((..(.(((((....)))))..)..))))))).)))....))))))))))))).((...((((....)))).-...))... ( -39.30) >DroYak_CAF1 35807 99 - 1 CCACACUGAUGCGGUCAAUGCGGUGGCAUUCAGUCGCGAUGGCGACAAGUUUGCCACCGGUGGCAGUGAUCGUCAGGUGAGUUU-UGUUUCUUGAUUG-------UU .(((.(((..(((((((.((((((((((..(.(((((....)))))..)..)))))))....))).))))))))))))).....-.............-------.. ( -39.00) >DroPer_CAF1 9688 103 - 1 CCAUACGGCGGCUGUAAAUGCUGUGGGCUUUUCCCAGGAUGGAGAGAAGUUUGCCACUGGUGGCAACGAUCGCCAGGUGAGUUGGGGCGGCUCAUCCA----AUGGC ((((..((((..(((..........((((((((((.....)).))))))))(((((....))))))))..)))).(((((((((...)))))))))..----)))). ( -40.00) >consensus CCACACUGAUGCGGUAAAUGCGGUGGCAUUCAGUCACGAUGGCGACAAGUUUGCCACCGGUGGCAGUGAUCGCCAGGUGAGUUUCUGUUUCUUAAUAA_GGUAUGAA .......((.((((.....((.(((((.....)))))..((((((.....((((((....))))))...)))))).....))..)))).))................ (-24.93 = -23.38 + -1.55)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:32:15 2006