
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 13,349,398 – 13,349,495 |
| Length | 97 |
| Max. P | 0.666385 |

| Location | 13,349,398 – 13,349,495 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.57 |
| Mean single sequence MFE | -31.92 |
| Consensus MFE | -22.49 |
| Energy contribution | -23.72 |
| Covariance contribution | 1.23 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.614093 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13349398 97 + 23771897 AAGGUCCUUCUGCAAGGAUCUCCAACCAUUUACCCCAUUUGGCUGCAAUGGGAUUG-----------------------CAUUACUUUGACUGCAGCAUGAUGCAGAGGUGGUCGGCACU .((((((((....))))))))((.(((((((..((((((.......)))))).(((-----------------------(((((..(((....)))..))))))))))))))).)).... ( -31.60) >DroSec_CAF1 20969 97 + 1 AAGGUCCUUCCGCAAGGAUCUCCAACCAUUUACCCCAUUUGGCUGCAAUGGGAUUG-----------------------CAUUACUUUGACUGCAGCAUGAUGCAGAGGUGGUCGGCGCU .((((((((....))))))))((.(((((((..((((((.......)))))).(((-----------------------(((((..(((....)))..))))))))))))))).)).... ( -31.60) >DroSim_CAF1 20789 97 + 1 AAGGUCCUUCCGCAAGGAUCUCCAACCAUUUACCCCAUUUGGCUGCAAUGGGAUUG-----------------------CAUUACUUUGACUGCAGCAUGAUGCAGAGGUGGCCGGCGCU ..((((..(((....)))......(((......((((((.......)))))).(((-----------------------(((((..(((....)))..)))))))).)))))))...... ( -29.00) >DroEre_CAF1 21316 95 + 1 AAGGUCCUUCCGCAAGGACCUCCAACCAUUUACCCCAUUUGGCUGCAAUGGGAUUG-----------------------CAUUACUUUGACUGCAGCAUGAUGCAGAGCCAGUCGGGC-- .((((((((....))))))))............(((..(((((((((((...))))-----------------------)..........(((((......)))))))))))..))).-- ( -34.20) >DroYak_CAF1 22077 97 + 1 AAGGUCCUUCCGCAAGGAUCUCCAACCAUUUACCCCAUUUGGCUGCAAUGGGAUUG-----------------------CAUUACUUUGACUGCAGCAUGGUGCAGAGGUGGUCGGUGUU .((((((((....))))))))((.(((((((..((((((.......)))))).(((-----------------------(((((..(((....)))..))))))))))))))).)).... ( -31.30) >DroAna_CAF1 19971 113 + 1 AGGGUCUUUCCACAGGGAUCUCCAGCCAUAAACUUCCAUUGGUUGCAAUGGGAUUGGAAUCCCCUUUCACCCGGGGAGGCAUUACUUUGACUACAGCAU---CCAGAGGA-CUCGGU--- .((((((((......((....)).(((......((((((((....))))))))......(((((........))))))))...................---...)))))-)))...--- ( -33.80) >consensus AAGGUCCUUCCGCAAGGAUCUCCAACCAUUUACCCCAUUUGGCUGCAAUGGGAUUG_______________________CAUUACUUUGACUGCAGCAUGAUGCAGAGGUGGUCGGCGCU .((((((((....))))))))((.(((((((..((((.(((....)))))))......................................(((((......)))))))))))).)).... (-22.49 = -23.72 + 1.23)
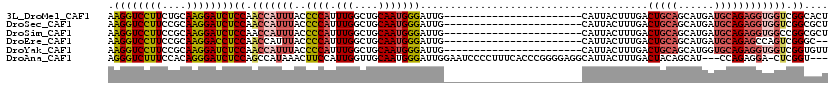
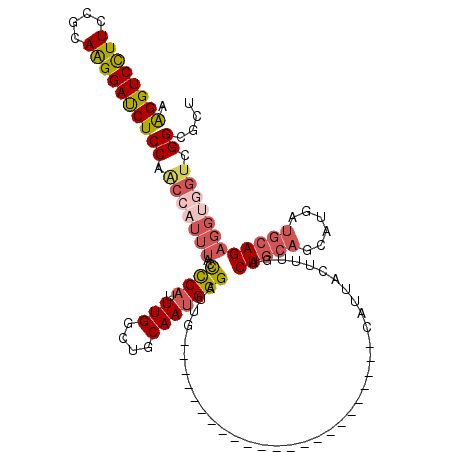
| Location | 13,349,398 – 13,349,495 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.57 |
| Mean single sequence MFE | -32.42 |
| Consensus MFE | -21.68 |
| Energy contribution | -23.38 |
| Covariance contribution | 1.70 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.666385 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 13349398 97 - 23771897 AGUGCCGACCACCUCUGCAUCAUGCUGCAGUCAAAGUAAUG-----------------------CAAUCCCAUUGCAGCCAAAUGGGGUAAAUGGUUGGAGAUCCUUGCAGAAGGACCUU ....(((((((....(((((..((((........)))))))-----------------------))((((((((.......))))))))...)))))))((.(((((....))))).)). ( -31.70) >DroSec_CAF1 20969 97 - 1 AGCGCCGACCACCUCUGCAUCAUGCUGCAGUCAAAGUAAUG-----------------------CAAUCCCAUUGCAGCCAAAUGGGGUAAAUGGUUGGAGAUCCUUGCGGAAGGACCUU ....(((((((....(((((..((((........)))))))-----------------------))((((((((.......))))))))...)))))))((.(((((....))))).)). ( -31.70) >DroSim_CAF1 20789 97 - 1 AGCGCCGGCCACCUCUGCAUCAUGCUGCAGUCAAAGUAAUG-----------------------CAAUCCCAUUGCAGCCAAAUGGGGUAAAUGGUUGGAGAUCCUUGCGGAAGGACCUU ....(((((((....(((((..((((........)))))))-----------------------))((((((((.......))))))))...)))))))((.(((((....))))).)). ( -31.20) >DroEre_CAF1 21316 95 - 1 --GCCCGACUGGCUCUGCAUCAUGCUGCAGUCAAAGUAAUG-----------------------CAAUCCCAUUGCAGCCAAAUGGGGUAAAUGGUUGGAGGUCCUUGCGGAAGGACCUU --.((((..((((((((((......))))).....((((((-----------------------......)))))))))))..))))...........(((((((((....))))))))) ( -35.80) >DroYak_CAF1 22077 97 - 1 AACACCGACCACCUCUGCACCAUGCUGCAGUCAAAGUAAUG-----------------------CAAUCCCAUUGCAGCCAAAUGGGGUAAAUGGUUGGAGAUCCUUGCGGAAGGACCUU ....(((((((....((((...((((........)))).))-----------------------))((((((((.......))))))))...)))))))((.(((((....))))).)). ( -30.60) >DroAna_CAF1 19971 113 - 1 ---ACCGAG-UCCUCUGG---AUGCUGUAGUCAAAGUAAUGCCUCCCCGGGUGAAAGGGGAUUCCAAUCCCAUUGCAACCAAUGGAAGUUUAUGGCUGGAGAUCCCUGUGGAAAGACCCU ---..(...-(((.(.((---((.((.((((((..........(((((........)))))........((((((....)))))).......)))))).))))))..).)))..)..... ( -33.50) >consensus AGCGCCGACCACCUCUGCAUCAUGCUGCAGUCAAAGUAAUG_______________________CAAUCCCAUUGCAGCCAAAUGGGGUAAAUGGUUGGAGAUCCUUGCGGAAGGACCUU ....(((((((...(((((......)))))....................................((((((((.......))))))))...)))))))((.(((((....))))).)). (-21.68 = -23.38 + 1.70)

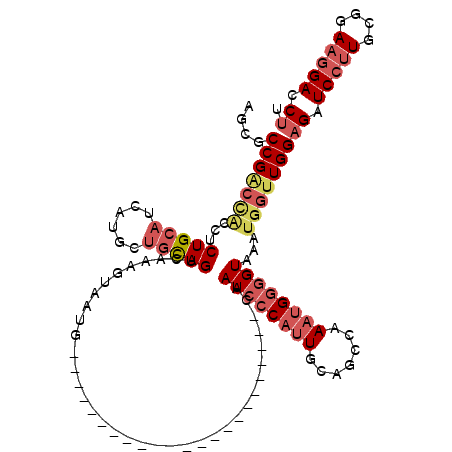

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:32:12 2006